Abstract
Amyotrophic lateral sclerosis (ALS) is the most common motor neuron diseases (MND), while frontotemporal lobar degeneration (FTLD) is the second most common cause of early-onset dementia. Many ALS families segregating FTLD have been reported, particularly over the last decade. Recently, mutations in TARDBP, FUS/TLS, and C9ORF72 have been identified in both ALS and FTLD patients, while mutations in VCP, a FTLD associated gene, have been found in ALS families. Distinct variants located in the 3′-untranslated region (UTR) of the SIGMAR1 gene were previously reported in three unrelated FTLD or FTLD–MND families. We directly sequenced the coding and UTR regions of the SIGMAR1 gene in a targeted cohort of 25 individual familial ALS cases of Caucasian origin with a history of cognitive impairments. This screening identified one variant in the 3′-UTR of the SIGMAR1 gene in one ALS patient, but the same variant was also observed in 1 out of 380 control chromosomes. Subsequently, we screened the same samples for a C9ORF72 repeat expansion: 52% of this cohort was found expanded, including the sample with the SIGMAR1 3′-UTR variant. Consequently, coding and noncoding variants located in the 3′-UTR region of the SIGMAR1 gene are not the cause of FTLD–MND in our cohort, and more than half of this targeted cohort is genetically explained by C9ORF72 repeat expansions.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common motor neuron diseases (MND), with an incidence of 1–2/100 000 person-years,1 while frontotemporal lobar degeneration (FTLD) is the second most common cause of early-onset dementia with an incidence of 3–5/100 000 person-years.2 Interestingly, up to 50% of ALS patients are deemed likely to develop some cognitive impairments.3 In 2006, the TDP-43 protein was found to be an important constituent of aggregates in neurons of MND–FTLD patients,4 which suggested a common pathological pathway for the two conditions. Mutations in the TARDBP gene encoding the TDP-43 protein,5, 6 and the FUS/TLS gene encoding the FUS protein7, 8 been identified in both ALS and FTLD patients, while mutations in ALS cases recently reported in VCP,9 a gene in which mutations were previously associated with FTLD.10 Furthermore, neurons of ALS and FTLD patients both display a nuclear clearing and cytoplasmic sequestration of normal cellular TDP-43 or FUS proteins,11 which are encoded by genes that, when mutated, are estimated to account for up to 5–10% of all ALS cases.6, 7 Twelve families linked to a locus on chromosome 9p have been reported to have members with either MND or FTLD, while a few members displayed both phenotypes.12, 13, 14, 15, 16, 17 The causative pathogenic hexanucleotide repeat in the gene C9ORF72 was recently identified by two different groups.18, 19 One of these reports found that this repeat expansion explains ∼3% of sporadic FTD, 11.7% of familial FTD, 4.1% of sporadic ALS, and 23.5% of familial ALS cases.18 The second report shows that the repeat expansion is detectable in about 50% of familial and 20% of sporadic Finnish cases, in more than one third of familial cases of wider European ancestry, and in more than 29% of the Finnish FTD population among which 36% have a family history.19 In addition, recently, a variant in the 3′-untranslated region (UTR) of the SIGMAR1 gene, encoding a receptor protein having an important role in various cellular functions, was reported to segregate in a FTLD–MND pedigree.20 Two additional 3′-UTR variants were identified by the same group in two unrelated FTLD families thought to be linked to the chromosome 9p locus, but no segregation could be observed in these. Another group recently conducted homozygosity mapping in a large consanguineous family with six members affected with juvenile ALS. They identified a SIGMAR1 missense mutation in all affected individuals.21 Considering the finding of variants in the SIGMAR1 gene and the recent publication of hexanucleotide repeat expansions in C9ORF72 located in the same chromosomal region, we wanted to evaluate the potential contribution of SIGMAR1 variants in ALS patients of European descents. We selected 25 individual familial ALS cases with a family history of cognitive impairments, and directly sequenced the entire coding and UTR regions of the SIGMAR1 gene. We also evaluated this specific targeted cohort for C9ORF72 repeat expansions.
Materials and Methods
Standard Protocol Approval, Registration, and Patient Consents
Protocols were approved by the ethics committee on human experimentation of the Center Hospitalier de l’Université de Montréal. All patients gave written informed consent after which patient information and blood were collected.
Subjects
Clinical information from 260 familial ALS cases of Caucasian origin not mutated in SOD1, TARDBP, or FUS was analyzed; 25 families were found to have a history of cognitive impairments. One affected member from each family was selected for sequencing the entire coding and UTR regions of the SIGMAR1 gene (NM_005866.2 ). In addition, 190 control participants were sequenced for one fragment encompassing the first 396 bp of the 3′-UTR, in which one novel substitution was identified in one familial ALS patient. Moreover, the same patients were assessed for the presence of expanded hexanucleotide repeat in C9ORF72 (NM_001256054.1).
Gene screening
Six sets of primers were used for each sample to amplify the open-reading frame of the SIGMAR1 gene (Table 1). The PCR products containing the exons included a minimum of 50 bp from each of the flanking introns. Primers were designed using the ExonPrimer software from the UCSC human genome browser website (http://genome.ucsc.edu/cgi-bin/hgGateway). Amplification was conducted by PCRs using the AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA, USA) as per manufacturer’s instructions. PCR products were amplified with the same procedure of direct sequencing in patients and controls, and were sequenced at the Genome Quebec Innovation Center (Montréal, Québec, Canada) using a 3730XL DNA analyzer (Montréal). Mutation survey or software (version 3.10) was used for mutation detection analyses (SoftGenetics, State College, PA, USA).
Hexanucleotide repeat analysis
We performed a repeat-primed PCR assay using the FastStart PCR Master Mix (Roche, Indianapolis, IN, USA) using the reported optimized conditions.18, 19 PCR products were analyzed on an ABI 3730 sequencer with GeneMapper software version 4.0 (Applied Biosystem, Carlsbad, CA, USA).
Results and Discussion
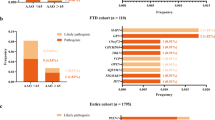
Only one variant (c.672* 43G>T) located in the 3′ UTR of the SIGMAR1 gene was identified in one patient. The substituted guanine is well conserved through different species. Other affected family members were not available for testing, but the variant was not present in six other unaffected family members. However, after testing 190 matched controls, the variant was identified in one control, suggesting that the substitution is not causative of ALS in the family. No other unknown variants were identified in the 25 patients tested. Interestingly, after performing the repeat-primed method, we found that 13 patients actually carried a hexanucleotide repeat expansion in C9ORF72. This represents 52% of the total tested. The cause of the disease for the remaining 12 patients is still unexplained genetically.
The SIGMAR1 gene was a positional candidate based on linkage in an Australian MND–FTLD family.17 The c.672*51G>T 3′-UTR substitution in the SIGMAR1 gene was identified in the linked family after the sequencing of about 200 candidate genes in the chromosome 9p candidate region. The nonpolymorphic nucleotide change segregated with the disease haplotype of this Australian pedigree. The variant was not identified in 1269 controls. In addition, c.672*26C>T and c.672*47G>A substitutions also located in the 3′-UTR of SIGMAR1 were identified in different FTLD pedigrees of Australian and Polish origins, respectively. These variants were also absent in matched controls. A modification in SIGMAR1 gene expression was found in the Australian-origin family, while a small expression difference was observed for the other two families, each with only one patient actually carrying the variant. It was also shown that modulating the expression of SIGMAR1 influences the cellular localization of TDP-43 and FUS, and it was suggested that the 3′-UTR of SIGMAR1 variant causes a pathogenic alteration of TARDBP and FUS expression levels.20 Considering that the C9ORF72 hexanucleotide repeat expansion was not reported when the SIGMAR1 mutations were identified and that the genes are close to each other on chromosome 9p, it is possible that the SIGMAR1 variants identified in the first report actually segregated with C9ORF72 expansions. The finding of a 3′-UTR variant in our specifically targeted cohort permitted to test and confirms this hypothesis.
Further studies in different populations will help to evaluate the contribution of the sigma nonopioid intracellular receptor 1 protein in MND and/or FTLD, and future studies need to address the reported effects on expression levels of the different proteins involved. However, based on our study, it can be concluded that coding mutations or UTR variants in SIGMAR1 are not a cause of ALS in our cohort of Caucasian ALS families with a history of cognitive impairments, but that repeat expansions in C9ORF72 genetically explain a significant proportion of the same cohort.
References
Johnston CA, Stanton BR, Turner MR et al. Amyotrophic lateral sclerosis in an urban setting: a population based study of inner city London. J Neurol 2006; 253: 1642–1643.
Mercy L, Hodges JR, Dawson K et al. Incidence of early-onset dementias in Cambridgeshire, United Kingdom. Neurology 2008; 71: 1496–1499.
Lillo P, Hodges JR : Frontotemporal dementia and motor neurone disease: overlapping clinic-pathological disorders. J Clin Neurosci 2009; 16: 1131–1135.
Neumann M, Sampathu DM, Kwong LK et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–133.
Benajiba L, Le Ber I, Camuzat A et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol 2009; 65: 470–473.
Kabashi E, Valdmanis PN, Dion P et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 2008; 40: 572–574.
Kwiatkowski TJ, Bosco DA, Leclerc AL et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009; 323: 1205–1208.
Broustal O, Camuzat A, Guillot-Noel L et al. FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J Alzheimers Dis 2010; 22: 765–769.
Johnson JO, Mandrioli J, Benatar M et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010; 68: 857–864.
Watts GD, Wymer J, Kovach MJ et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 2004; 36: 377–381.
Lagier-Tourenne C, Cleveland DW : Rethinking ALS: the FUS about TDP-43. Cell 2009; 136: 1001–1004.
Vance C, Al-Chalabi A, Ruddy D et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain 2006; 129: 868–876.
Morita M, Al-Chalabi A, Andersen PM et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology 2006; 66: 839–844.
Valdmanis PN, Dupre N, Bouchard JP et al. Three families with amyotrophic lateral sclerosis and frontotemporal dementia with evidence of linkage to chromosome 9p. Arch Neurol 2007; 64: 240–245.
Le Ber I, Camuzat A, Berger E et al. Chromosome 9p-linked families with frontotemporal dementia associated with motor neuron disease. Neurology 2009; 72: 1669–1676.
Boxer AL, Mackenzie IR, Boeve BF et al. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatry 2011; 82: 196–203.
Luty AA, Kwok JB, Thompson EM et al. Pedigree with frontotemporal lobar degeneration--motor neuron disease and Tar DNA binding protein-43 positive neuropathology: genetic linkage to chromosome 9. BMC Neurol 2008; 8: 32.
Dejesus-Hernandez M, Mackenzie IR, Boeve BF et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–256.
Renton AE, Majounie E, Waite A et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72: 257–268.
Luty AA, Kwok JB, Dobson-Stone C et al. Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration-motor neuron disease. Ann Neurol 2010; 68: 639–649.
Al-Saif A, Al-Mohanna F, Bohlega S : A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol 2011; 70: 913–919.
Acknowledgements
VVB and HD are supported by the Canadian Institutes of Health Research. We would like to thank the patients and the families involved in this study, and to acknowledge support from the Association pour la Recherche sur la Sclérose Latérale Amyotrophique (ARS), the Association Française contre les Myopathies (AFM), and the French Group on MND. GAR holds the Canada’s Research Chair in Neurogenetics and a Jeanne-et-J.-Louis-Levesque Chair for the Genetics of Brain Diseases.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Belzil, V., Daoud, H., Camu, W. et al. Genetic analysis of SIGMAR1 as a cause of familial ALS with dementia. Eur J Hum Genet 21, 237–239 (2013). https://doi.org/10.1038/ejhg.2012.135
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.135
Keywords
This article is cited by
-
Genotype-phenotype correlations of amyotrophic lateral sclerosis
Translational Neurodegeneration (2016)
-
Genotype-phenotype relationship in hereditary amyotrophic lateral sclerosis
Translational Neurodegeneration (2015)
-
Gene panels and primers for next generation sequencing studies on neurodegenerative disorders
Molecular & Cellular Toxicology (2015)
-
In silico analysis of SIGMAR1 variant (rs4879809) segregating in a consanguineous Pakistani family showing amyotrophic lateral sclerosis without frontotemporal lobar dementia
neurogenetics (2015)