Abstract
Both receptor-interacting protein kinase 1 (RIPK1) and RIPK3 can signal cell death following death receptor ligation. To study the requirements for RIPK-triggered cell death in the absence of death receptor signaling, we engineered inducible versions of RIPK1 and RIPK3 that can be activated by dimerization with the antibiotic coumermycin. In the absence of TNF or other death ligands, expression and dimerization of RIPK1 was sufficient to cause cell death by caspase- or RIPK3-dependent mechanisms. Dimerized RIPK3 induced cell death by an MLKL-dependent mechanism but, surprisingly, also induced death mediated by FADD, caspase 8 and RIPK1. Catalytically active RIPK3 kinase domains were essential for MLKL-dependent but not for caspase 8-dependent death. When RIPK1 or RIPK3 proteins were dimerized, the mode of cell death was determined by the availability of downstream molecules such as FADD, caspase 8 and MLKL. These observations imply that rather than a ‘switch’ operating between the two modes of cell death, the final mechanism depends on levels of the respective signaling and effector proteins.
Similar content being viewed by others
Main
Mammalian cells can use a number of mechanisms to kill themselves. The best characterized depends on the Bcl-2 family members Bax and Bak that work via mitochondria to activate caspases.1 Some caspases, notably caspase 8, can be activated independently of Bcl-2 family members, for example, after stimulation of members of the TNF receptor superfamily.2 Recently, it has become apparent that some of these receptors, including TNFR1, can activate a third suicide mechanism that does not require caspases, and in which the morphology of the dying cell differs from classical apoptosis. This form of cell death, termed ‘necroptosis’, can often be blocked by necrostatin-1 (nec-1), an inhibitor of the kinase activity of receptor-interacting protein kinase 1 (RIPK1).3, 4 Accordingly, observations from several groups have shown that in some cell types, expression of RIPK1 can signal cell death by caspase-independent necroptosis.5
It has previously been revealed that RIPK1 could function downstream of death receptors, but in those cases, cell death was usually blocked by coexpression of the viral inhibitor of caspases 1 and 8, CrmA,6 and typically exhibited a classical ‘apoptotic’ morphology. It was revealed that RIPK1 engages FADD via homotypic binding of their death domains (DDs), and FADD in turn activates caspase 8.6, 7
RIPK3, like RIPK1, bears a kinase domain and RIP homology interaction motif (RHIM), but unlike RIPK1 does not have a DD.8, 9, 10, 11 RIPK3 is required for necroptosis.12, 13 Furthermore, RIPK1 appears to activate RIPK3 in this pathway, as cell death could be blocked by nec-1.14 RIPK3 activates, by phosphorylation, MLKL, a pseudokinase essential for this death pathway.15, 16, 17 Once activated, MLKL forms multimers that trigger breaches of the plasma membrane.18, 19, 20
Although RIPK3 is necessary for necroptosis, it is unclear whether activation of RIPK3 is sufficient for cell death, because TNF activates signaling by many pathways in addition to those controlled by RIPK1.21 It is also unclear whether RIPK3 can contribute to apoptosis. Despite some reports to this effect,8, 9, 22 RIPK3 has been described as the necroptotic ‘switch’, implying its activity precipitates necroptosis to the exclusion of apoptosis.23, 24, 25
Here, we have directly activated RIP kinases without the confounding effects of multiple signals emanating from the target cell’s cytokine receptors, allowing us to define more precisely the functions of RIPK1 and RIPK3. We activated RIP kinases by dimerization using inducible lentiviral vectors, each encoding a chimera of a RIP kinase with subunit B of E. coli DNA gyrase.26 We infected mouse embryonic fibroblasts (MEFs) that lack genes for, or expression of, various cell death proteins, induced expression of the RIPK chimera, caused its dimerization by addition of the divalent antibiotic coumermycin (C) and quantitated the resulting cell death.
Our results reveal that each of RIPK1 and RIPK3 can contribute to both apoptosis and necroptosis depending on the biochemical context. Furthermore, necroptosis can occur in the absence of caspase 8 and FADD, which shows that the ripoptosome, with core components caspase 8, FADD and RIPK1,27, 28 is not required for necroptosis. Instead, we propose that dimers of RIPK1 and/or RIPK3 are the pivotal complexes from which both forms of cell death can progress.
Results
Dimerization of RIPK1 or RIPK3 is sufficient to cause death of MEFs
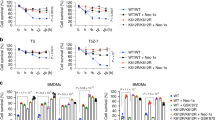
We generated lentiviral constructs that express chimeric RIPK1 gyrase and RIPK3 gyrase fusion proteins when induced with 4 hydroxytamoxifen (4HT) (Figure 1a). Induction of RIPK1 gyrase (Figure 1b) was sufficient to induce death of wild-type (WT) MEFs, and the number of cells that died was greatly increased by dimerization after the addition of coumermycin (Figure 1c). In this experiment, 4HT induction of RIPK3 gyrase (Figure 1b) did not lead to measurable cell death until coumermycin was added to induce dimerization (Figure 1c). Therefore, expression and dimerization of RIPK1 or RIPK3 is sufficient to induce cell death without adding a further stimulus, such as TNF.
Expression and dimerization of RIPK1 gyrase and RIPK3 gyrase is sufficient to cause death of MEFs. (a) FLAG-tagged RIPK1 gyrase B and FLAG-tagged RIPK3 gyrase B constructs. Inducible expression is driven by binding of a GAL4-T2 estrogen Receptor-VP16 transcription factor to 5 upstream UAS elements in the presence of 4HT.50 The divalent antibiotic coumermycin (C) dimerizes the gyrase B domains. (b) Western blots of lysates from MEFs bearing the inducible RIPK1 gyrase or RIPK3 gyrase constructs in the absence or presence (+) of 16 nM 4HT for 6 h were probed with antibodies to RIPK1 or RIPK3. The same blots were probed with antibodies to β-actin as a loading control, either simultaneously, in the case of the RIPK1 blot, or sequentially in the case of the RIPK3 blot. Endogenous and induced fusion proteins are labeled. (c) Analysis of cell viability by propidium iodide exclusion in untreated cells, and those treated for 24 h with 800 nM C alone, 16 nM 4HT alone or 4HT+C. PI-negative (viable) cells are boxed, and their percentages are indicated. Although induction of RIPK1 gyrase alone caused some cell death (depending on expression levels), addition of coumermycin greatly increased the death of cells expressing either the RIPK1 gyrase or RIPK3 gyrase proteins
Expression of death-inducing proteins is frequently lost in immortalized cell lines
To determine the consequences of dimerizing the RIP kinases, we not only induced the constructs in a variety of MEF lines derived from gene-deleted embryos, but we also took advantage of the fact that several immortalized cell lines have varying levels of death-inducing proteins. As shown in Figure 2a, MLKL expression was found to be low in the Fadd−/− and one Ripk1−/− MEF line (3.6); RIPK3 levels were low in Ripk1−/− MEFs 1.7 and 2.6 and undetectable in caspase 8−/− MEFs; and MLKL was particularly high in our subline of L929 fibroblastoid cells.
MEF lines and L929 cells express varying amounts of cell death proteins. (a) Lysates were prepared from WT and gene-deleted MEF lines as well as L929 mouse fibroblastoid cells, and identical aliquots were run on three replicate gels and, after blotting, were probed with antibodies to the indicated proteins. The gene-deleted lines lacked expression of the corresponding proteins, but, in addition, the Fadd−/− MEFs expressed barely detectable levels of MLKL, the Ripk1−/− line 3.6 expressed very low levels of MLKL, the Ripk3−/− line had low levels of RIPK1 and the caspase 8−/− line did not express detectable levels of RIPK3. (b) TNF alone causes death of L929 cells, strongly inhibited by nec-1 but unaffected by QVD. Death of Mlkl−/− MEFs caused by dimerization of RIPK3 gyrase is strongly inhibited by QVD but not by nec-1. L929 cells were treated with 100 ng/ml human Fc-TNF and Mlkl−/− cells were treated with 16 nM 4HT and 800 nM C for 24 h in the presence or absence of 50 μM nec-1 or 10 μM QVD. Viability was determined by PI exclusion on a flow cytometer. Triangles and circles indicate independently performed experiments, and error bars show S.E.M. values
L929 cells have an unusual phenotype: they are predisposed to necroptosis, for example, when treated with a combination of TNF and caspase inhibitors.29 Furthermore, a subline of L929 has been described that does not require caspase inhibition for this response.14 Figure 2b shows that the L929 subline used in our laboratory falls into this latter category, given that treatment with the pan-caspase inhibitor Q-VD-OPh (QVD)30 failed to prevent death induced by TNF in L929, even though the same concentrations of QVD were able to protect RIPK3 gyrase-triggered death of Mlkl−/− MEFs (Figure 2b and see below). In contrast, the L929 cells can be protected from TNF-induced killing by nec-1, as expected for a necroptotic death induced in this manner. Thus, despite bearing substantial levels of caspase 8 (Figure 2a), this subline of L929 is clearly resistant to caspase 8-dependent apoptosis. This phenotype of L929 cells has been attributed to inadequate recruitment of caspase 8 to FADD,31 thus rendering the cells effectively deficient in available caspase 8.
Dimerized RIPK1 induces death via both caspase 8-dependent and RIPK3-dependent pathways
RIPK1 is known to act in both caspase-dependent (apoptotic) and RIPK3–MLKL-dependent (necroptotic) death pathways.32 To confirm which effectors were required downstream of RIPK1 for each death mechanism, we expressed RIPK1 gyrase in either WT MEFs or those lacking components of the death pathways. When RIPK1 gyrase was induced and dimerized in various lines of MEFs (Figure 3a), death occurred, except in the caspase 8−/− line that also lacked RIPK3 (Figure 3b). To determine how much of the RIPK1 gyrase-induced death in the susceptible cell lines required caspase activity, we added QVD. In most cell types, QVD greatly reduced, but did not completely block, RIPK1 gyrase-induced death, indicating that more than half of the dying cells were killed by apoptosis. However, in the cells lacking RIPK3, QVD gave complete protection against dimerized RIPK1 gyrase, suggesting that when RIPK3 is absent, RIPK1 can only induce caspase-dependent death, but when RIPK3 is present, RIPK1 can also trigger a caspase-independent death mechanism.
Dimerization of RIPK1 can induce death of L929 cells by a caspase-independent mechanism and death of MEFs by both caspase-dependent and caspase-independent mechanisms. (a) Various MEF lines and L929 cells bearing the inducible RIPK1 gyrase construct were either untreated or treated with 4HT for 6 h. Lysates were separated by SDS-PAGE, blotted and probed with antibodies to RIPK1 and β-actin as a loading control. Low MW bands in lanes from treated cells are assumed to be breakdown products of the RIPK1 gyrase resulting from incomplete inhibition of proteases during cell lysis. (b) Fibroblast lines bearing the inducible RIPK1 gyrase construct were either untreated or treated with 4HT, 4HT+C or 4HT+C after pretreatment for 1 h with 10 μM QVD to inhibit caspase activity. After a further 24 h, viability of the cells was determined by PI exclusion. Mean±S.E.M. are shown for three independent sets of experiments. The levels of endogenous RIPK3 and caspase 8 (as determined by western blot; see Figure 2a) in each line are indicated below the histograms. The mechanism of cell death was inferred from the protection given by caspase inhibition with QVD, and caspase-independent cell death was presumed to be necroptosis. (c) In L929 cells, death induced by expression of RIPK1 gyrase was not inhibited by QVD, but was reduced by nec-1, and hence is mainly necroptotic. When RIPK1 gyrase was dimerized by coumermycin, inhibition of cell death by nec-1 was much less effective. The percentage of PI-negative (viable) cells is shown in the boxes
Consistent with the inability of our L929 cells to undergo caspase 8-dependent apoptosis, pretreatment with QVD did not prevent death induced by RIPK1 gyrase (Figure 3b), but death was substantially lessened by addition of nec-1 (Figure 3c, right-hand panels). Furthermore, this protection afforded by nec-1 was reduced from 38 to 10% when RIPK1 gyrase was dimerized with coumermycin (Figure 3c, lower versus upper right panels), raising the possibility that enforced dimerization interferes with or overcomes the conformational perturbation that underlies inhibition of RIPK1 kinase activity by nec-1.
A summary of the activity of dimerized RIPK1 is shown in Figure 3b. Wherever functional caspase 8 was available, caspase-dependent apoptosis occurred; the presence of RIPK3 allowed caspase-independent necroptosis, and where both were present, both death mechanisms occurred, although not necessarily in the same cell. Where neither RIPK3 nor caspase 8 was present, no death occurred.
In the absence of caspase 8 or FADD, dimerized RIPK3 gyrase induces only MLKL-dependent cell death
To see which proteins were required for death induced by dimerized RIPK3 gyrase, we induced it in MEFs lacking expression of other components of the cell death mechanisms, as we had for RIPK1 gyrase. Expression and dimerization of RIPK3 gyrase in caspase 8−/− MEFs (Figure 4a) killed ∼70% of the cells within 24 h and, as expected, this death was not preventable by QVD (Figure 4b).
RIPK3 gyrase can cause cell death by a mechanism that requires neither caspase 8 nor RIPK1 (necroptosis). (a) Western blot analysis of lysates from caspase 8−/− MEFs bearing the inducible RIPK3 gyrase construct in the absence or presence (+) of 16 nM 4HT for 6 h. The same blots were probed with antibodies to RIPK3, and reprobed with antibodies to β-actin as a loading control. (b) Caspase 8−/− MEFs bearing the inducible FLAG-RIPK3 gyrase construct were either untreated or treated with combinations of 16 nM 4HT and 800 nM C, with or without 10 μM QVD, for 24 h. Viability of the cells was determined by PI exclusion. Mean±S.E.M. of five independently performed experiments are shown. Predictably, QVD was unable to prevent death of the caspase 8−/− MEFs, indicating that RIPK3 was activating another death mechanism, presumably necroptosis. (c) Western blot of lysates of L929 cells bearing the inducible RIPK3 gyrase construct in the absence or presence (+) of 16 nM 4HT for 6 h. The same blots were probed with antibodies to RIPK3, and reprobed with antibodies to β-actin as a loading control. (d) L929 cells bearing the inducible FLAG-RIPK3 gyrase construct were either untreated or treated with combinations of 16 nM 4HT, 800 nM C, 10 μM QVD and 100 ng/ml TNF with or without 50 μM nec-1 for 24 h, and cell viability was determined by PI exclusion. Mean±S.E.M. of five independently performed experiments are shown. Dimerized RIPK3 caused death of the L929 cells, and this was not inhibited by QVD, or by the same doses of nec-1 that were able to block necroptosis induced by TNF. (e) Western blot analysis of lysates from Fadd−/− MEFs bearing a 4HT-inducible RIPK3 gyrase construct and a doxycycline (dox)-inducible MLKL construct in the absence or presence (+) of 16 nM 4HT with or without 100 ng/ml dox for 6 h. The same blot was probed sequentially with antibodies to RIPK3, MLKL and β-actin as a loading control. Note that these FADD −/− MEFs do not express detectable levels of endogenous MLKL. (f) Fadd−/− MEFs were either untreated or treated with combinations of 16 nM 4HT, 800 nM C and 100 ng/ml dox, with or without 10 μM QVD, for 24 h. Viability was determined by PI exclusion. Mean±S.E.M. of six independently performed experiments are shown. Dimerized RIPK3 was not able to cause death of the cells unless MLKL was also induced, and the death that occurred was not inhibited by QVD, and was therefore presumably necroptosis. (g) Western blot analysis of lysates from a Ripk1−/− MEF line (3.6) bearing a 4HT-inducible RIPK3 gyrase construct and a dox-inducible MLKL construct in the absence or presence (+) of 16 nM 4HT with or without 100 ng/ml dox for 6 h. The same blot was probed sequentially with antibodies to MLKL, RIPK3 and β-actin as a loading control. (h) Ripk1−/− 3.6 MEFs were either untreated or treated with combinations of 16 nM 4HT, 800 nM C and 100 ng/ml dox, with or without 10 μM QVD, for 24 h. Viability was determined by PI exclusion. Mean±S.E.M. of three independently performed experiments are shown. Dimerized RIPK3 was able to cause some death of the Ripk1−/− cells, whereas MLKL alone had no effect. When MLKL and RIPK3 dimers were induced together, more cell death occurred, and this was not blocked by QVD
Induction and dimerization of RIPK3 gyrase (Figure 4c) caused ∼90% of L929 cells to die (Figure 4d). Their death was not reduced by nec-1, consistent with RIPK3 acting downstream of RIPK1 (Figure 4d). As with their death caused by RIPK1 gyrase (Figure 3b), and consistent with the inability of caspase 8 to activate in L929 cells, their death caused by RIPK3 gyrase was not inhibited by QVD.
As shown previously,33 the Fadd−/− MEFs we used expressed very low levels of MLKL (Figure 2a). Induction and dimerization of RIPK3 gyrase (Figure 4e) failed to induce death (Figure 4f). However, coexpression of MLKL (via a dox-inducible vector) restored the ability of RIPK3 gyrase to trigger death of the Fadd−/− cells, and, as with the caspase 8−/− cells, this death was not inhibited by QVD (Figure 4f). These experiments show that MLKL is required for RIPK3 to trigger necroptosis and, because induction of MLKL alone did not cause cell death, in order to kill, exogenously expressed MLKL, and presumably endogenous WT MLKL, must first be activated by RIPK3.
In the absence of RIPK1, RIPK3 mainly causes MLKL-dependent death
Western blots revealed that MEFs derived from various Ripk1−/− embryos expressed varying amounts of MLKL (Figure 2a). In one line in which MLKL was barely detectable (Ripk1−/− 3.6), induction and dimerization of RIPK3 gyrase (Figure 4g) caused negligible cell death (Figure 4h). However, as for the Fadd−/− cells, restoration of MLKL allowed RIPK3 gyrase to cause death that was not preventable by QVD (Figure 4h).
Together, these data confirm that FADD and caspase 8 are not required for RIPK3 to trigger necroptosis, and that RIPK3 acts downstream of RIPK1 in the necroptotic pathway.
Dimerized RIPK3 can also induce cell death by a caspase 8-dependent mechanism
Figure 5a shows induction of RIPK3 gyrase in WT and Ripk1−/− 1.7 MEFs that express endogenous MLKL. Figure 5b shows the effects of the induced and dimerized proteins. There were two major differences between the responses of the WT and Ripk1−/− cells. First, more WT than Ripk1−/− cells were killed by RIPK3 gyrase (77% WT MEFs dead as opposed to 50% Ripk1−/− MEFs) and, second, although QVD did not protect either cell type completely, the effect was greater, and only reached statistical significance, on the WT cells (P=10−4 for WT versus 0.07 for Ripk1−/−). These results imply that in addition to the necroptotic mechanism demonstrated above, RIPK3 can activate a second pathway that depends on caspase(s) and partly on RIPK1.
Activation of RIPK3 can trigger caspase-dependent apoptosis as well as MLKL-dependent necroptosis. (a) Western blot analysis of lysates from WT and Ripk1−/− 1.7 MEF lines bearing a 4HT-inducible RIPK3 gyrase construct in the absence or presence (+) of 16 nM 4HT for 6 h. The same blots were probed with antibodies to RIPK3 and reprobed with antibodies to β-actin as a loading control. (b) WT (dark gray columns; data from three independent cell lines) and Ripk1−/− MEFs (light gray columns; data combined from lines 1.7 and 2.6) were either untreated or treated with combinations of 16 nM 4HT, 800 nM C, 50 μM nec-1 and/or 10 μM QVD for 24 h. Viability was determined by PI exclusion. Mean±S.E.M. of 3–5 independently performed experiments are shown. RIPK3 was able to cause death of both WT and Ripk1−/− cells, but it caused more cell death in the presence of RIPK1. In neither case was the death affected by nec-1, but it was reduced by QVD, indicating that RIPK3 can trigger a caspase-dependent death process, but it does so more efficiently in cells expressing RIPK1. (c) Western blot analysis of lysates from Mlkl−/− MEFs bearing a 4HT-inducible RIPK3 gyrase construct in the absence or presence (+) of 16 nM 4HT for 6 h. The same blots were probed with antibodies to RIPK3 and reprobed with antibodies to β-actin as a loading control. (d) Mlkl−/− MEFs were either untreated or treated with combinations of 16 nM 4HT and 800 nM C, with or without 50 μM nec-1 and/or 10 μM QVD, for 24 h. Viability was determined by PI exclusion. Mean±S.E.M. of nine independently performed experiments are shown. RIPK3 was able to cause death of Mlkl−/− cells that was completely blocked by QVD, indicating that in the absence of MLKL, activated RIPK3 causes only caspase-dependent cell death. (e) WT and Ripk1−/− MEFs bearing the RIPK3 gyrase construct were treated with 10 nM 4HT for 24 h and/or 700 nM C for 2 h. Lysates were run on replicate blots and were probed with antibodies to full-length caspase 8, processed caspase 8, processed caspase 3 and PARP. The full-length caspase 8 blots were washed and re-probed for β-actin to act as a loading control. (f) Western blot analysis of lysates from L929 cells and WT MEF lines bearing a 4HT-inducible RIPK3 gyrase construct in the absence or presence (+) of 16 nM 4HT with or without 800 nM C for 5 h. The same blots were probed sequentially with antibodies to PARP, pro-caspase 8 and β-actin as a loading control. Dimerized RIPK3 gyrase can activate caspase 8 (but not detectably in L929 cells) and lead to processing of downstream caspases and PARP, and it does so more efficiently in the presence of RIPK1
In the absence of MLKL, RIPK3 induces only apoptosis
Figure 5c shows induction of RIPK3 gyrase in Mlkl−/− MEFs, and Figure 5d shows the effect of the induced and dimerized chimeric protein. The cells were efficiently killed by dimerized RIPK3, and fully protected by QVD, confirming that dimerized RIPK3 is capable of activating an apoptotic process independent of MLKL.
RIPK3 can induce processing of caspases 8 and 3, and this is more efficient if RIPK1 is present
Induction and dimerization of RIPK3 gyrase in WT MEFs was followed by robust cleavage of caspase 8, caspase 3 and PARP (Figures 5e and f), consistent with the ability of RIPK3 to activate a caspase-dependent apoptotic mechanism. L929 cells again served as a negative control; just as we had been unable to detect apoptosis (Figure 4d), we were unable to detect PARP cleavage (Figure 5f).
Surprisingly, in the Ripk1−/− MEFs we detected processing of caspases 8 and 3 in response to RIPK3 dimerization, but it was at lower levels than in the WT cells, and did not result in detectable PARP cleavage at this time point (2 h after addition of coumermycin). These results show that dimerized RIPK3 gyrase can activate caspase 8 (Figure 5e) and cause apoptosis (Figures 5b and d), but it is less efficient in the absence of RIPK1 (Figures 4h, 5b and e). Although unexpected, this ability of RIPK3 to partially trigger caspase 8 processing even in the absence of RIPK1 is consistent with our previous observations that when RIPK3 levels are elevated, TNF can trigger processing of caspase 8 in the absence of RIPK1.33
Activation of RIPK3 fails to induce death when both MLKL and caspase 8 are absent
To determine whether dimerization of RIPK3 could induce cell death by any other mechanism, in addition to the caspase and MLKL pathways, we expressed the RIPK3 gyrase vector in caspase 8−/−/Mlkl−/− mouse dermal fibroblast (MDF) cells (Figure 6a). Induction and dimerization caused no death of the caspase 8−/−/Mlkl−/− cells, whereas in contrast, caspase8−/−/Ripk3−/− cells died by necroptosis that, as expected, was not blocked by QVD (Figure 6b).
In the absence of both caspase 8 and MLKL, activated RIPK3 fails to induce death. (a) Western blot analysis of lysates from WT, caspase 8−/−Ripk3−/− and caspase 8−/−Mlkl−/− mouse dermal fibroblasts (MDFs) either untreated or treated (+) with 10 nM 4HT for 24 h. The blots were probed for FLAG followed by β-actin as a loading control. (b) WT, caspase 8−/−Ripk3−/− and caspase 8−/−Mlkl−/− MDFs were either untreated or treated with combinations of 10 nM 4HT, 700 nM C and 10 μM QVD for 24 h. Viability was determined by PI exclusion. Mean±S.E.M. of three independently performed experiments are shown. Dimerized RIPK3 was able to cause some death of the WT cells, and this could be partly blocked by QVD. When both caspase 8 and MLKL were absent, no death was induced
RIPK3-induced necroptosis is dependent on the catalytic activity of its kinase domain
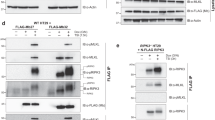
RIPK1 kinase activity has been shown to be necessary for necroptosis induced by the FasL, TNF and TRAIL death receptors,5, 34 but its requirement for caspase-dependent apoptosis is controversial.6, 31, 35 To test whether the kinase activity of RIPK3 was required for either form of RIPK3 gyrase-induced death, we initially generated a K51A kinase-dead mutant of the RIPK3 gyrase vector.9 We expressed it in WT MEFs, immunoprecipitated the RIPK3 gyrase and confirmed that the kinase was inactive in an in vitro assay (Figure 7a). The K51A RIPK3 gyrase protein did not cause death of caspase 8−/− cells (Figure 7b) that are susceptible to necroptosis (Figure 4b) but lack detectable expression of endogenous RIPK3 (Figure 2a). However, the K51A construct appeared to trigger a small number of WT and Ripk3−/− cells to die, and this was blocked by QVD (Figure 7b). No death was detectable in the apoptosis-susceptible Mlkl−/− cells (Figure 7b), nor was cleavage of PARP in WT cells (Figure 7c), but these may both be explained by lower levels of the K51A mutant protein compared with WT RIPK3 gyrase (Figures 7c and d, track 6).
K51A mutant of RIPK3 gyrase is catalytically inactive but apparently unstable. (a) WT and K51A RIPK3 gyrase were immunoprecipitated using anti-FLAG coupled beads from cells expressing the respective constructs, and used in in vitro kinase assays to show whether they were able to auto-phosphorylate. Washed beads were resuspended in 30 μl of kinase buffer (20 mM HEPES, pH 7.5, 10 mM MnCl2 and 1 mM DTT), divided into two aliquots that were incubated in the presence versus absence of 250 μM ATP at 30°C for 20 min and analyzed by electrophoresis followed by replicate western blots that were probed with anti-phosphothreonine and anti-RIPK3. (b) WT, Mlkl−/−, Ripk3−/−, caspase 8−/− MEFs and L929 cells bearing WT or K51A (kinase dead) RIPK3 gyrase constructs were either untreated or treated with combinations of 16 nM 4HT, 800 nM C and 10 μM QVD for 16 h. Viability was determined by PI exclusion. Mean±S.E.M. of three independently performed experiments are shown. The kinase-mutant K51A RIPK3 was unable to cause statistically significant levels of cell death in the apoptosis-prone Mlkl−/−, or in caspase 8−/− cells (that lack detectable RIPK3). It appeared to induce death of a small number of WT and Ripk3−/− cells, but it was only able to cause convincing levels of cell death in L929 cells that express endogenous RIPK3 and show reduced activation of caspase 8. The death in L929 cells could not be blocked by QVD, and hence it is presumed to be necroptosis. Note the low levels of expression of K51A relative to WT in (a), (c) and track 6 of (d). (c) Western blots of lysates from MEFs bearing WT or K51A (kinase dead) RIPK3 gyrase constructs that were untreated or treated with 16 nM 4HT and 700 nM C for 6 h were probed sequentially with antibodies to caspase 8, PARP and RIPK3, followed by β-actin as loading controls. The asterisk indicates a nonspecific band. (d) Western blots of lysates from WT, Mlkl−/−, Ripk3−/−, caspase 8−/− MEFs and L929 cells bearing the K51A (kinase dead) RIPK3 gyrase construct, and WT and L929 cells bearing the WT RIPK3 gyrase were untreated or treated with 16 nM 4HT for 6 h and probed with antibodies to RIPK3 followed by β-actin as loading controls
Given the inconclusive nature of these results, and the difficulty in attaining levels of expression of the K51A mutant comparable to those of WT (see also Figure 7a), we examined two further RIPK3 mutants predicted to have compromised kinase activity. In the RIPK3 catalytic domain, we substituted asparagine for the crucial catalytic residue, an aspartic acid at position 143 (D143N).36 A third mutant, R142G, had been identified in a colon carcinoma specimen.37 Correct geometry of the HRD catalytic motif (at positions 141–143) is imperative for kinase activity.38 Thus, we predicted that mutation of the HRD motif to HGD in the R142G RIPK3 mutant would lead to conformational pliability within the catalytic loop and concomitant defective catalytic activity.
Purified recombinant WT, R142G and D143N RIPK3 kinase domains (Figure 8a) were subjected to in vitro kinase assays using 32P-γ-ATP and the pseudokinase domain of mouse MLKL as substrates. At comparable protein concentrations (Figure 8b, left panel), the WT protein robustly phosphorylated both itself and MLKL, the D143N mutant entirely lacked detectable catalytic activity, whereas that of the R142G mutant was markedly reduced (Figure 8b, right panel). We prepared RIPK3 gyrase vectors containing each of the two mutations, and introduced these constructs into cell lines. Like the K51A mutant (Figure 7b) when induced and dimerized in caspase 8−/− MEFs (Figure 8c), which are susceptible to necroptosis (Figure 4b) but lack expression of endogenous RIPK3 (Figure 2a), no statistically significant death was caused by the kinase-deficient (R142G) or kinase-dead (D143N) RIPK3 gyrase proteins (Figure 8d). In contrast, the WT construct robustly caused cell death that, as expected, was not blocked by QVD. These results indicate that in the absence of endogenous RIPK3, and even when expression levels are comparable with those of the WT form, kinase-inactive or -compromised RIPK3 gyrase is unable to induce necroptosis.
The kinase activity of RIPK3 is needed for it to cause necrosis but not apoptosis. (a and b) Recombinant D143N and R142G mutant mRIPK3 kinase domains are catalytically defective. (a) A total volume of 1 μg of wild-type (WT), D143N and R142G mRIPK3 kinase domain, which had been expressed in insect cells and purified, was resolved by reducing SDS-PAGE and stained with Coomassie Blue. Comparable intensity of Coomassie-stained bands confirmed reliability of concentration estimation by Absorbance at 280 nm. These concentration estimates were used subsequently to ensure that comparable quantities of WT and mutant RIPK3 were assayed in in vitro kinase assays. (b) In vitro kinase assays demonstrate that WT, but not D143N, mRIPK3 can autophosphorylate and phosphorylate recombinant MLKL, consistent with our prior characterization of recombinant D143N mRIPK3,16 whereas R142G mRIPK3 exhibits defective catalytic activity in autophosphorylation and MLKL phosphorylation. The shown experiment is representative of two independent experiments. BSA was added as a carrier protein (100 ng/μl). The Coomassie-stained image of the representative gel is shown on the left and the autoradiograph of the same gel on the right. Only 25 ng of each recombinant RIPK3 kinase domain was loaded in each experiment and is consequently not visible by Coomassie Blue staining. (c) Western blots of lysates from MEFs bearing WT, D143N (kinase dead) and R142G (kinase compromised) RIPK3 gyrase constructs that were untreated or treated with 16 nM 4HT and 800 nM C for 6 h were probed sequentially with antibodies to PARP and RIPK3, followed by β-actin as loading controls. The asterisks indicate nonspecific bands. Although a background level of cleaved PARP is detectable in all cell types in this experiment, the band intensity increases with induction of expression and dimerization of RIPK3 gyrase only in Ripk3−/− and Mlkl−/− cells, in which caspase-dependent apoptosis occurs (see f and g). (d–g) Kinase-inactive or -compromised RIPK3 gyrase can induce apoptosis, but cannot induce necroptosis without the presence of (endogenous) WT RIPK3. (d) Caspase 8−/− cells bearing the inducible RIPK3 gyrase construct were either untreated or treated with combinations of 16 nM 4HT, 800 nM C and 10 μM QVD. Mean±S.E.M. of four independently performed experiments are shown. (e) In the presence of WT RIPK3, kinase-inactive RIPK3 gyrase induces necroptosis. L929 cells bearing the inducible RIPK3-gyrase construct were either untreated or treated as in (d), or with the addition of nec-1. Mean±S.E.M. of four independently performed experiments are shown. The R142G mutant was as effective as the WT, and even the D143N mutant induced some cell death. (f) Apoptosis induced by dimerized RIPK3 is independent of its kinase activity. Mlkl−/− cells bearing the respective RIPK3 gyrase constructs were untreated or treated as in (e). Mean±S.E.M. of three independently performed experiments are shown. Both R142G and D143N mutants caused the death of substantial numbers of cells, almost equivalent to WT. (g) Apoptosis induced by dimerized RIPK3 is independent of bystander RIPK3. Ripk3−/− MEFs bearing the respective RIPK3 gyrase constructs were untreated or treated as in (e). Mean±S.E.M. of three independently performed experiments are shown. Again, cell death was substantial in response to the WT and each of the mutants
Kinase-dead and kinase-compromised RIPK3 gyrase can cause necroptosis in the presence of endogenous WT RIPK3
In L929 cells that express abundant endogenous RIPK3 (Figure 2a), induction and dimerization of the kinase-compromised RIPK3 gyrase mutant (R142G) killed as many cells as the WT protein (Figure 8e). Furthermore, the kinase-dead RIPK3 gyrase constructs K51A and D143N were also able to trigger some death (Figures 7b and 8e). A possible explanation for the ability of the kinase mutant forms of RIPK3 to trigger necroptosis in L929 cells but not in MEFs lacking expression of endogenous RIPK3 (i.e., caspase 8−/− cells) is that although kinase-dead RIPK3 gyrase is unable to activate MLKL directly, it may be able to recruit endogenous RIPK3 that will then auto-activate to form a partially active necroptotic complex. In this case, the inability of kinase-dead RIPK3 to induce death of WT MEFs treated with QVD might be because of lower levels of endogenous RIPK3 or MLKL compared with those in L929 cells.
Dimerization also activates the nonkinase function of RIPK3 to induce apoptosis
Both D143N and R142G mutant forms of RIPK3 gyrase caused substantial death in apoptosis-susceptible (Mlkl−/−) cells (Figure 8f), and, consistent with this, triggered proteolytic cleavage of PARP (Figure 8c). Thus, we conclude that although kinase activity of RIPK3 is required for necroptosis, it is not necessary to trigger apoptosis. Consistent with this, expression of the mutants in Ripk3−/− cells caused PARP cleavage and apoptosis that did not require the presence of endogenous RIPK3 (Figures 8c and g). Furthermore, as apoptosis induced by these RIPK3 mutants was not blocked by nec-1, for RIPK3 to induce apoptosis, it also does not require the kinase activity of RIPK1 (Figures 8f and g). Finally, as for RIPK1 gyrase (Figure 3b, Ripk3−/− cells), dimerization of RIPK3 gyrase greatly increased the extent of apoptosis (Figures 8f and g), in which its kinase activity was not involved. Therefore, homodimerization of RIPK3 not only serves to activate its kinase activity, but also serves a kinase-independent function in triggering activation of FADD and caspase 8 to cause apoptosis.
Discussion
Forced homodimerization has been used previously to directly activate kinases and caspases by facilitating their autophosphorylation and autocleavage respectively.26, 39, 40 Here we have shown that dimerization of RIPK1 or RIPK3 can induce kinase-dependent necroptosis, provided that active RIPK3 and MLKL are present. Importantly, at least in the case of dimerized RIPK3, the kinase activity can be supplied by bystander endogenous RIPK3 monomers that are presumably recruited to a kinase-inactive dimer. Furthermore, necroptosis can be induced in this manner in the absence of FADD or caspase 8, and therefore independently of the so-called ripoptosome.27, 28
In some molecular contexts, forced dimerization of one protein can induce dimerization, and thereby activation, of interacting proteins, such as when death receptors activate caspase 8 to induce apoptosis.41 Accordingly, we have shown that when FADD and caspase 8 were present, dimerization of RIPK1 or RIPK3 induced apoptosis that was completely independent of the kinase activity of the dimeric RIPK as well as of any bystander endogenous forms. Thus, in the context of the apoptotic machinery (the ripoptosome) it appears that, without activating the kinase, dimerization of the RIPKs induces apoptosis, probably via the C terminal domain.8, 9, 22
If dimerization of the two different RIPK domains has different outcomes, what determines the mode of cell death? This is most clearly answered by examining the death of Ripk1−/− cells following dimerization of RIPK3. When MLKL was absent, RIPK3 was able to trigger death of a small number of cells by apoptosis (Figure 4h); in the absence of RIPK1 and the presence of moderate levels of MLKL, RIPK3 triggered both caspase-dependent and -independent death (Figure 5b); when high levels of MLKL were present, cells died by necroptosis (Figure 4h). Thus, both activation of RIPK1 and of RIPK3 are capable of inducing cell death by both apoptotic and necroptotic mechanisms, but which mode predominates depends on the levels of the respective downstream effector proteins. These results are depicted in Figures 9a and b.
RIPK dimers are at the fork in the cell death pathway. (a) Depending on the availability of targets, forcibly dimerized RIPK1 recruits FADD and caspase 8 to induce apoptosis, or if autophosphorylation occurs, it recruits and phosphorylates RIPK3 that in turn phosphorylates MLKL to induce necroptosis. There is a slight bias toward apoptosis. (b) Similarly, forcibly dimerized RIPK3 recruits FADD and therefore caspase 8, but this is inefficient unless RIPK1 is available. If autophosphorylation occurs in the RIPK3 dimer, it phosphorylates MLKL and necroptosis follows. We predict that in normal cells responding to death signals, RHIM mediated homo- and hetero-dimerization of RIPK1 and RIPK3 mimics these activities. In the case of RIPK1, the dimerization can be via its DD
Our findings are supported by a report by Newton et al.,42 published during revision, in which a knock-in mouse bearing another kinase-inactive RIPK3 mutant (D161N) was characterized. Consistent with our data, that study demonstrated that RIPK3 catalytic activity is dispensable for apoptosis, but essential for necroptosis. However, in contrast to that study,42 we observed that apoptosis could proceed, to some extent, in the absence of RIPK1.
Our observations prompt consideration of an alternative map of the pathways to cell death following an extrinsic signal such as TNF. Given that forced dimerization of either RIPK1 or RIPK3 can mimic RHIM-mediated heterodimerization in inducing autophosphorylation and necroptosis, we predict that RHIM-mediated homo- or heterodimerization can also, like forced homodimerization, indirectly activate FADD and caspase 8, leading to apoptosis. In the case of RIPK1, this could also be achieved by DD-mediated dimerization. Then, the outcomes would be similar to those depicted in Figure 9. If the first effect of dimerization is activation of the kinases, and RIPK3 is either part of the initial dimer or available to be recruited, and MLKL is available, then necroptosis ensues. If, instead, the dimer recruits FADD before kinase activation takes place, and caspase 8 is available, the apoptotic cascade begins. The presence of RIPK1, either in the initial dimer or as a secondary recruit, increases the efficiency of induction of apoptosis. Feedback regulation by caspase 8,22, 43, 44 and possibly by other kinases,45 will refine the outcome, but in principle, apoptosis and necroptosis need not be mutually exclusive and could proceed simultaneously.
Materials and Methods
Genetically modified cell lines and mice
Production of MEF lines has been described previously in detail.46 Briefly, primary MEFs were generated from E10–15 embryos and then infected with SV40 Large T antigen expressing lentivirus to generate immortal cell lines. Ripk1+/− mice were provided by Michelle Kelliher (University of Massachusetts Medical School, Worcester, MA, USA) and Ripk3 −/− mice were provided by Kim Newton (Genentech Inc., Oceanside, CA, USA). Fadd−/− MEFs were provided by Francis Chan (University of Massachusetts Medical School), Mlkl−/− MEFs were provided by Warren Alexander, and the L929 cell line was from Andreas Strasser (both at the Walter and Eliza Hall Institute, Melbourne, VIC, Australia). Caspase 8 gene-deleted MEFs were generated from E8.5 caspase 8 LoxP/LoxP embryos from mice provided by Dr. Stephen Hedrick (UCSD, San Diego, CA, USA) as previously described.33 Caspase 8−/−/Mlkl−/− and caspase 8−/−/Ripk3−/− MDFs were generated as previously described.47
Antibodies and chemicals
Primary antibodies used for western blot analysis were anti-FLAG (F-3165, Sigma, Sydney, NSW, Australia), anti-β-actin (A-1978, Sigma), anti-RIPK1 (610458, BD Transduction Laboratories, North Ryde, NSW, Australia), anti-RIPK3 (Pharmingen, North Ryde, NSW, Australia; 551042), anti-caspase 8 (Lorraine O’Reilly, The Walter and Eliza Hall Institute), anti-cleaved caspase 8 (Asp 387) (8592, Cell Signaling Technology, Danvers, MA, USA), anti-cleaved caspase 3 (Asp 175) (9661, Cell Signaling Technology), anti-PARP (9542, Cell Signaling Technology), anti-MLKL (3H1, Millipore MABC604, Billerica, MA, USA; provided by Jian-Guo Zhang, The Walter and Eliza Hall Institute), anti-FADD (clone IF7-05486, Millipore) and anti-Phospho-Threonine (Cell Signaling Technology). 4HT, coumermycin and doxycycline were purchased from Sigma, and Q-VD-OPh (OPH001) was from SM Biochemicals (Anaheim, CA, USA). Necrostatin-1 was obtained from Sigma and Dr. Mark McKinlay (TetraLogic Pharmaceuticals, Malvern, PA, USA).
Recombinant protein expression and purification
cDNAs encoding Mus musculus MLKL (179–464)16 and WT or mutant Mus musculus RIPK3 kinase domain were cloned into the pFastBac HTb vector (Life Technologies, Mulgrave, VIC, Australia). Mutations in the mRIPK3 kinase domain were introduced by oligonucleotide-directed mutagenesis PCR. All inserts were verified by Sanger sequencing (Micromon, Monash University, Melbourne, Australia).
Bacmids and baculoviruses were generated using established procedures,48, 49 before recombinant proteins were expressed and purified from Sf21 insect cells essentially as previously described.16, 48 Briefly, following cell lysis by sonication, N-terminal His6-tagged proteins were purified by Ni2+-affinity chromatography and the tag cleaved by incubation with TEV protease for 2 h at 22°C. Detagged mMLKL (179–464) was extensively dialyzed, before further Ni2+ chromatography to eliminate undigested protein and TEV protease followed by a final Superdex-200 gel filtration (GE Healthcare, Rydalmere, NSW, Australia) in 200 mM NaCl and 20 mM HEPES, pH 7.5. TEV-digested WT and mutant mRIPK3 kinase domains were concentrated by centrifugal ultrafiltration before resolution by Superdex-200 gel filtration chromatography in 200 mM NaCl, 20 mM Tris, pH 8, 10% v/v glycerol and 0.5 mM TCEP. Fractions containing mRIPK3 kinase domain were pooled and subjected to ion-exchange (MonoQ, GE Healthcare) chromatography with a linear gradient from buffer A (20 mM Tris, pH 8, 10% v/v glycerol and 0.5 mM TCEP) to buffer B (0.5 M NaCl, 20 mM Tris, pH 8, 10% v/v glycerol and 0.5 mM TCEP) over 20 column volumes. Finally, fractions containing purified proteins were pooled, concentrated by centrifugal ultrafiltration to 3–6 mg/ml, aliquoted and snap frozen before storage at −80°C until required.
In vitro kinase assays on purified proteins
Kinase assays for purified recombinant WT, R142G and D143N kinase domains were performed as described previously.16 Briefly, 100 ng of recombinant WT or mutant mRIPK3 kinase domain was incubated for 5 min at 25°C in the presence of 3.5 μg mMLKL (179–464) after initiation of the kinase reaction by the addition of 5 μCi 32P-γ-ATP and cold ATP to 37.5 μM in a 40 μl reaction. Kinase reactions were performed in kinase reaction buffer: 100 mM NaCl, 20 mM Tris, pH 8, 4 mM MgCl2, 0.1 mM DTT and 0.1 mg/ml BSA. Reactions were terminated by boiling in 1 × reducing sample buffer for 5 min before resolution on a 4–12% Bis-Tris SDS-PAGE gel. The gel was Coomassie Blue stained, dried and exposed to a phosphor screen for 16 h and read by Phosphorimager (Fujifilm, Campbellfield, VIC, Australia).
Generation of RIPK1 and RIPK3 gyrase constructs
A fusion construct of mouse FLAG-RIPK1 or FLAG-RIPK3 and a fragment of E. coli gyrase B (residues 2–220) that dimerizes in response to the antibiotic coumermycin26 was cloned into the 4HT inducible lentiviral vector pF 5xUAS46, 50 and confirmed by Sanger sequencing. Mutations within the mRIPK3 kinase domains (K51A, D143N and R142G) were subcloned into the FLAG-RIPK3 gyrase construct in place of the WT sequence and verified by Sanger sequencing (AGRF, Walter and Eliza Hall Institute and Micromon Facility, Monash University).
Cell culture, transfections and lentiviral infections
Cell lines were maintained at 37°C, 10% CO2 in DMEM supplemented with 10% (v/v) fetal bovine serum (Gibco, Melbourne, VIC, Australia), 50 μg/ml penicillin G, 50 U/ml streptomycin and 2 mM L-glutamine. SV40 Large T antigen was cloned into the lentiviral vector pFU as described previously.46 Lentiviruses were generated by transfecting subconfluent 10 cm plates of 293T cells with vector plasmids together with the packaging constructs pCMV-ΔR8 and pVSV-G using Effectene (Qiagen, Chadstone Centre, VIC, Australia) as described previously.50 After 48 h, viral supernatants were collected, filtered, supplemented with 4 μg/ml polybrene, added to the target MEFs and centrifuged at 2800 r.p.m. for 90 mins at 28°C. After a recovery period of 48 h, stably infected cells were selected in the presence of 5 μg/ml puromycin and 250 μg/ml hygromycin B. Expression of pF 5xUAS inducible constructs was induced with 16 nM 4HT and dimerization of the FLAG-RIPK gyrase B chimeric protein was induced by 800 nM coumermycin unless otherwise indicated. Mouse MLKL was expressed from the doxycycline-inducible pF TRE3G PGK puro vector as previously described.33
Cell death assays
Cells were seeded at 67 000 cells per well into 12-well tissue culture plates. Where indicated, 16 nM 4HT and/or 800 nM coumermycin or 10 μM Q-VD-OPh were added to cells for 24 h and cell death measured by uptake of propidium iodide (PI) using a FACScalibur flow cytometer (BD Biosciences, North Ryde, NSW, Australia). A total of 10 000 events per sample were collected, and the percentage of live cells (% PI-negative cells) quantified using WEASEL software (version 2.2.2, Walter and Eliza Hall Institute).
Western blotting
Lysates were prepared in DISC lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol and 1% Triton X-100), supplemented with protease inhibitor cocktail (Roche, Castle Hill, NSW, Australia) and clarified by centrifugation. Protein samples were separated on 4–12% polyacrylamide gels (Invitrogen, Mulgrave, VIC, Australia) and transferred to PVDF membrane (Immobilon, Merck, Kilsyth, VIC, Australia) for incubation with specified antibodies. Membrane blocking steps and antibody dilutions were performed using 5% (v/v) skim milk or 2% BSA in Tris-buffered saline containing 0.1% (v/v) Tween-20 (TBST), and washing steps performed with TBST. Western blots were visualized by enhanced chemiluminescence (GE, Rydalmere, NSW, Australia).
Immunoprecipitation and in vitro kinase assay of immunoprecipitates
Immunoprecipitations were performed using standard procedures as previously described.51 In vitro kinase assays were performed as previously described52 on the immunoprecipitates, except that unlabeled ATP was used. Samples were separated on 4–12% PAGE gels, transferred to PVDF membranes and probed using anti-phosphothreonine antibodies.
Abbreviations
- RIPK1:
-
receptor-interacting protein kinase 1
- nec-1:
-
necrostatin-1
- DD:
-
death domain
- WT:
-
wild type
- RHIM:
-
RIP homology interaction motif
- MEF:
-
mouse embryonic fibroblast
- 4HT:
-
4 hydroxytamoxifen
- MDF:
-
mouse dermal fibroblast
References
Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997; 91: 479–489.
Wang L, Du F, Wang X . TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703.
Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 2005; 1: 112–119.
Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G . Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714.
Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 2000; 1: 489–495.
Grimm S, Stanger BZ, Leder P . RIP and FADD: two “death domain”-containing proteins can induce apoptosis by convergent, but dissociable, pathways. Proc Natl Acad Sci USA 1996; 93: 10923–10927.
Micheau O, Tschopp J . Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003; 114: 181–190.
Sun X, Lee J, Navas T, Baldwin DT, Stewart TA, Dixit VM . RIP3, a novel apoptosis-inducing kinase. J Biol Chem 1999; 274: 16871–16875.
Yu PW, Huang BC, Shen M, Quast J, Chan E, Xu X et al. Identification of RIP3, a RIP-like kinase that activates apoptosis and NFkappaB. Curr Biol 1999; 9: 539–542.
Kasof GM, Prosser JC, Liu D, Lorenzi MV, Gomes BC . The RIP-like kinase, RIP3, induces apoptosis and NF-kappaB nuclear translocation and localizes to mitochondria. FEBS Lett 2000; 473: 285–291.
Pazdernik NJ, Donner DB, Goebl MG, Harrington MA . Mouse receptor interacting protein 3 does not contain a caspase-recruiting or a death domain but induces apoptosis and activates NF-kappaB. Mol Cell Biol 1999; 19: 6500–6508.
He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 1100–1111.
Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372.
Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 2011; 35: 908–918.
Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227.
Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 2013; 39: 443–453.
Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA 2012; 109: 5322–5327.
Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol 2014; 16: 55–65.
Chen X, Li W, Ren J, Huang D, He WT, Song Y et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res 2014; 24: 105–121.
Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell 2014; 54: 133–146.
Cabal-Hierro L, Lazo PS . Signal transduction by tumor necrosis factor receptors. Cell Sig 2012; 24: 1297–1305.
Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N et al. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Sig 2007; 19: 2056–2067.
Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336.
Vandenabeele P, Melino G . The flick of a switch: which death program to choose? Cell Death Diff 2012; 19: 1093–1095.
Moriwaki K, Chan FK . RIP3: a molecular switch for necrosis and inflammation. Genes Dev 2013; 27: 1640–1649.
Farrar MA, Alberol-Ila J, Perlmutter RM . Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature 1996; 383: 178–181.
Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43: 432–448.
Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463.
Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W et al. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med 1998; 187: 1477–1485.
Caserta TM, Smith AN, Gultice AD, Reedy MA, Brown TL . Q-VD-OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis 2003; 8: 345–352.
Duprez L, Bertrand MJ, Vanden Berghe T, Dondelinger Y, Festjens N, Vandenabeele P . Intermediate domain of receptor-interacting protein kinase 1 (RIPK1) determines switch between necroptosis and RIPK1 kinase-dependent apoptosis. J Biol Chem 2012; 287: 14863–14872.
Vanlangenakker N, Vanden Berghe T, Vandenabeele P . Many stimuli pull the necrotic trigger, an overview. Cell Death Diff 2012; 19: 75–86.
Moujalled DM, Cook WD, Okamoto T, Murphy J, Lawlor KE, Vince JE et al. TNF can activate RIPK3 and cause programmed necrosis in the absence of RIPK1. Cell Death Dis 2013; 4: e465.
Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123.
Kim JW, Choi EJ, Joe CO . Activation of death-inducing signaling complex (DISC) by pro-apoptotic C-terminal fragment of RIP. Oncogene 2000; 19: 4491–4499.
Hanks SK, Quinn AM, Hunter T . The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 1988; 241: 42–52.
Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet 2008 Chapter 10, Unit 10.11.
Kornev AP, Haste NM, Taylor SS, Eyck LF . Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA 2006; 103: 17783–17788.
Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM . An induced proximity model for caspase-8 activation. J Biol Chem 1998; 273: 2926–2930.
Oberst A, Pop C, Tremblay AG, Blais V, Denault JB, Salvesen GS et al. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J Biol Chem 2010; 285: 16632–16642.
Spencer DM, Belshaw PJ, Chen L, Ho SN, Randazzo F, Crabtree GR et al. Functional analysis of Fas signaling in vivo using synthetic inducers of dimerization. Curr Biol 1996; 6: 839–847.
Newton K, Dugger DL, Wickliffe KE, Kapoor N, Cristina de-Almagro M, Vucic D et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 2014; 343: 1357–1360.
Lin Y, Devin A, Rodriguez Y, Liu ZG . Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev 1999; 13: 2514–2526.
O'Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R et al. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol 2011; 13: 1437–1442.
McQuade T, Cho Y, Chan FK . Positive and negative phosphorylation regulates RIP1- and RIP3-induced programmed necrosis. Biochem J 2013; 456: 409–415.
Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 2007; 131: 682–693.
Etemadi N, Holien JK, Chau D, Dewson G, Murphy JM, Alexander WS et al. Lymphotoxin alpha induces apoptosis, necroptosis and inflammatory signals with the same potency as tumour necrosis factor. FEBS J 2013; 280: 5283–5297.
Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA et al. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J 2014; 457: 323–334.
Babon JJ, Murphy JM . In vitro JAK kinase activity and inhibition assays. Methods Mol Biol 2013; 967: 39–55.
Callus BA, Ekert PG, Heraud JE, Jabbour AM, Kotevski A, Vince JE et al. Cytoplasmic p53 is not required for PUMA-induced apoptosis. Cell Death Diff 2008; 15: 213–215.
Vince JE, Chau D, Callus B, Wong WW, Hawkins CJ, Schneider P et al. TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1-TRAF2 complex to sensitize tumor cells to TNFalpha. J Cell Biol 2008; 182: 171–184.
Lock P, Abram CL, Gibson T, Courtneidge SA . A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J 1998; 17: 4346–4357.
Acknowledgements
This work was funded by NHMRC Grants 1016701 (to DLV) and 1057905 (to JMM) and Fellowship 1020136 (to DLV), and Cancer Council Victoria Grant 1044722 (to DLV and WDC), Cure Cancer Australia Foundation Grant 1050301 (to DMM), a Center Grant from Leukemia and Lymphoma Society (US) 7413-07, an ARC Future Fellowship FT100100100 (to JMM), and was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS (361646). We thank Toru Okamoto for the doxycycline-inducible vector, Francis Chan for the Fadd−/− MEFs, Kim Newton and Vishva Dixit for Ripk3−/− mice and Stephen Hedrick for caspase 8 conditional knockout mice.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Rights and permissions
About this article
Cite this article
Cook, W., Moujalled, D., Ralph, T. et al. RIPK1- and RIPK3-induced cell death mode is determined by target availability. Cell Death Differ 21, 1600–1612 (2014). https://doi.org/10.1038/cdd.2014.70
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.70
This article is cited by
-
Saracatinib inhibits necroptosis and ameliorates psoriatic inflammation by targeting MLKL
Cell Death & Disease (2024)
-
OASL phase condensation induces amyloid-like fibrillation of RIPK3 to promote virus-induced necroptosis
Nature Cell Biology (2023)
-
Repurposing of Ibrutinib and Quizartinib as potent inhibitors of necroptosis
Communications Biology (2023)
-
PRMT5-mediated regulatory arginine methylation of RIPK3
Cell Death Discovery (2023)
-
The Lck inhibitor, AMG-47a, blocks necroptosis and implicates RIPK1 in signalling downstream of MLKL
Cell Death & Disease (2022)