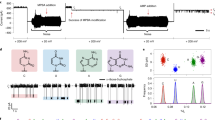
Abstract
RNA and DNA strands produce ionic current signatures when driven through an α-hemolysin channel by an applied voltage. Here we combine this nanopore detector with a support vector machine (SVM) to analyze DNA hairpin molecules on the millisecond time scale. Measurable properties include duplex stem length, base pair mismatches, and loop length. This nanopore instrument can discriminate between individual DNA hairpins that differ by one base pair or by one nucleotide.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rief, M. Clausen-Schaumann, H. & Gaub, H.E. Sequence-dependent mechanics of single DNA molecules. Nat. Struct. Biol. 6, 346–349 (1999).
Perkins, T.T., Quake, S.R., Smith, D.E. & Chu, S. Relaxation of a single DNA molecule observed by optical microscopy. Science 264, 822–826 (1994).
Smith, S.B., Cui, Y. & Bustamante, C. Overstretching B-DNA: the elastic response of individual double- stranded and single-stranded DNA molecules. Science 271, 795–799 (1996).
Voss, D. Gene express. New Sci. 164, 40–43 (1999).
Akeson, M., Branton, D., Kasianowicz, J.J., Brandin, E. & Deamer, D.W. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 77, 3227–3233 (1999).
Meller, A., Nivon, L., Brandin, E., Golovchenko, J. & Branton, D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. USA 97, 1079–1084 (2000).
Kasianowicz, J.J., Brandin, E., Branton, D. & Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 93, 13770–13773 (1996).
Bayley, H., Braha, O. & Gu, L.Q. Stochastic sensing with protein pores. Advan. Mater. 12, 139–142 (2000).
Song, L. et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore [see comments]. Science 274, 1859–1866 (1996).
Senior, M.M., Jones, R.A. & Breslauer, K.J. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc. Natl. Acad. Sci. USA 85, 6242–6246 (1988).
Michael, D. Chem-Site 3.01. (Pyramid Learning LLC, Hudson, OH; 1999).
Vapnik, V. The nature of statistical learning theory, Edn. 2. (Springer-Verlag, New York; 1999).
Burges, C.J.C. A tutorial on Support Vector Machines for pattern recognition. Data Mining and Knowledge Discovery 2, 121–167 (1998).
Cormen, T.H., Leiserson, C.E. & Rivest, R.L. Introduction to algorithms. (McGraw-Hill, New York; 1989).
Nievergelt, Y. Wavelets made easy. (Birkhauser, Boston; 1999).
Jaakkola, T.S. & Haussler, D. Exploiting generative models in discriminative classifiers. In Advances in neural information processing systems 11. (eds Kearnes, M.S., Solla, S.A. & Cohn, D.A.) (MIT Press, Cambridge, MA; 1999).
SantaLucia, J. Jr., A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95, 1460–1465 (1998).
Acknowledgements
This work was supported by NRHGI grant HG01826-01. We wish to thank Laura Steinmann for thoughtful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vercoutere, W., Winters-Hilt, S., Olsen, H. et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat Biotechnol 19, 248–252 (2001). https://doi.org/10.1038/85696
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/85696
This article is cited by
-
Implementation of biomolecular logic gate using DNA and electrically doped GaAs nano-pore: a first principle paradigm
Journal of Molecular Modeling (2021)
-
Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore
Nature Communications (2018)
-
Discriminating single-bacterial shape using low-aspect-ratio pores
Scientific Reports (2017)
-
Single-molecule analysis of lead(II)-binding aptamer conformational changes in an α-hemolysin nanopore, and sensitive detection of lead(II)
Microchimica Acta (2016)
-
α-Hemolysin nanopore studies reveal strong interactions between biogenic polyamines and DNA hairpins
Microchimica Acta (2016)