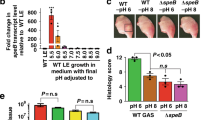
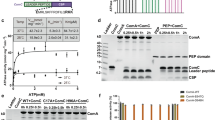
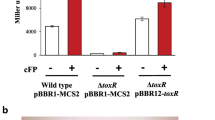
Abstract
Bacteria of the genus Enterococcus are the main causes of highly antibiotic-resistant infections that are acquired in hospitals1,2. Many clinical isolates of Enterococcus faecalis produce an exotoxin called cytolysin that contributes to bacterial virulence3. In addition to its toxin activity, the cytolysin is bactericidal for nearly all Gram-positive organisms4. An understanding of conditions that regulate cytolysin expression has advanced little since its initial description5. Here we show that the products of two genes, cylR1 and cylR2, which lack homologues of known function, work together to repress transcription of cytolysin genes. Derepression occurs at a specific cell density when one of the cytolysin subunits reaches an extracellular threshold concentration. These observations form the basis of a model for the autoinduction of the cytolysin by a quorum-sensing mechanism involving a two-component regulatory system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992–April 2000, issued June 2000. Am. J. Infect. Contr. 28, 429–448 (2000).
Mundy, L. M., Sahm, D. F. & Gilmore, M. Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13, 513–522 (2000).
Haas, W. & Gilmore, M. S. Molecular nature of a novel bacterial toxin: the cytolysin of Enterococcus faecalis. Med. Microbiol. Immunol. (Berl) 187, 183–190 (1999).
Brock, T. D., Peacher, B. & Pierson, D. Survey of the bacteriocines of enterococci. J. Bacteriol. 86, 702–707 (1963).
Todd, E. W. A comparative serological study of streptolysins derived from human and from animal infections, with notes on pneumococcal hemolyson, tetanolysin and staphylococcus toxin. J. Pathol. Bacteriol. 39, 299–321 (1934).
Gilmore, M. S., Segarra, R. A. & Booth, M. C. An HlyB-type function is required for expression of the Enterococcus faecalis hemolysin/bacteriocin. Infect. Immun. 58, 3914–3923 (1990).
Booth, M. C. et al. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel antibiotic. Mol. Microbiol. 21, 1175–1184 (1996).
Coburn, P. S., Hancock, L. E., Booth, M. C. & Gilmore, M. S. A novel means of self-protection, unrelated to toxin activation, confers immunity to the bactericidal effects of the Enterococcus faecalis cytolysin. Infect. Immun. 67, 3339–3347 (1999).
Poyart, C. & Trieu-Cuot, P. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to β-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156, 193–198 (1997).
Cserzo, M., Wallin, E., Simon, I., von Heijne, G. & Elofsson, A. Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10, 673–676 (1997).
Hofmann, K. & Stoffel, W. TMbase–a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374, 166 (1993).
Tusnady, G. E. & Simon, I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283, 489–506 (1998).
Brennan, R. G. & Matthews, B. W. The helix-turn-helix DNA binding motif. J. Biol. Chem. 264, 1903–1906 (1989).
Schultz, J., Milpetz, F., Bork, P. & Ponting, C. P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA 95, 5857–5864 (1998).
Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 28, 263–266 (2000).
Gilmore, M. S. et al. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 176, 7335–7344 (1994).
Dunny, G. M. & Winans, S. C. Cell–Cell Signaling in Bacteria (ASM, Washington DC, 1999).
Ike, Y. & Clewell, D. B. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J. Bacteriol. 158, 777–783 (1984).
Hoch, J. A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3, 165–170 (2000).
Zhang, H. Z., Hackbarth, C. J., Chansky, K. M. & Chambers, H. F. A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science 291, 1962–1965 (2001).
Francia, M. V. et al. Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid 46, 117–127 (2001).
Dong, Y. H. et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411, 813–817 (2001).
Sambrook, J. & Russell, D. W. Molecular Cloning (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001).
Shepard, B. D. & Gilmore, M. S. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl. Environ. Microbiol. 65, 1470–1476 (1999).
Acknowledgements
We thank P. Coburn, L. Hancock, M. Engelbert, C. Cox, K. Hatter, D. Johnson, B. Jett and K. Gilmore for technical support and critical review of the manuscript. This work was supported by grants from the National Institutes of Health and Research to Prevent Blindness.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Haas, W., Shepard, B. & Gilmore, M. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415, 84–87 (2002). https://doi.org/10.1038/415084a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/415084a
This article is cited by
-
The pathogenesis of prevalent aerobic bacteria in aerobic vaginitis and adverse pregnancy outcomes: a narrative review
Reproductive Health (2022)
-
Regulation of Enterococcus faecalis Biofilm Formation and Quorum Sensing Related Virulence Factors with Ultra-low Dose Reactive Species Produced by Plasma Activated Water
Plasma Chemistry and Plasma Processing (2019)
-
Lactobacillus plantarum isolated from cheese: production and partial characterization of bacteriocin B391
Annals of Microbiology (2017)
-
One-pot synthesis of class II lanthipeptide bovicin HJ50 via an engineered lanthipeptide synthetase
Scientific Reports (2016)
-
Bioluminescence based biosensors for quantitative detection of enterococcal peptide–pheromone activity reveal inter-strain telesensing in vivo during polymicrobial systemic infection
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.