Abstract
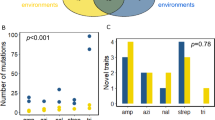
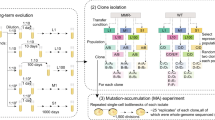
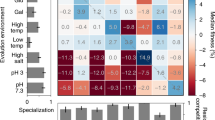
When organisms adapt genetically to one environment, they may lose fitness in other environments1,2,3,4. Two distinct population genetic processes can produce ecological specialization—mutation accumulation and antagonistic pleiotropy5,6,7,8. In mutation accumulation, mutations become fixed by genetic drift in genes that are not maintained by selection; adaptation to one environment and loss of adaptation to another are caused by different mutations. Antagonistic pleiotropy arises from trade-offs, such that the same mutations that are beneficial in one environment are detrimental in another. In general, it is difficult to distinguish between these processes5,6,7,8. We analysed the decay of unused catabolic functions in 12 lines of Escherichia coli propagated on glucose for 20,000 generations9,10. During that time, several lines evolved high mutation rates11. If mutation accumulation is important, their unused functions should decay more than the other lines, but no significant difference was observed. Moreover, most catabolic losses occurred early in the experiment when beneficial mutations were being rapidly fixed, a pattern predicted by antagonistic pleiotropy. Thus, antagonistic pleiotropy appears more important than mutation accumulation for the decay of unused catabolic functions in these populations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mills, D. R., Peterson, R. L. & Spiegelman, S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Natl Acad. Sci. USA 58 , 217–224 (1967).
Futuyma, D. J. & Moreno, G. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19, 207– 233 (1988).
Fry, J. D. Tradeoffs in fitness on different hosts: evidence from a selection experiment with a phytophagous mite. Am. Nat. 136, 569–580 (1990).
Bennett, A. F. & Lenski, R. E. Evolutionary adaptation to temperature. II. Thermal niches of experimental lines of Escherichia coli. Evolution 47, 1–12 ( 1993).
Rose, M. R. & Charlesworth, B. A test of evolutionary theories of senescence. Nature 287, 141– 142 (1980).
Rose, M. R. Evolutionary Biology of Aging (Oxford Univ. Press, Oxford, 1991).
Holt, R. D. Demographic constraints in evolution: towards unifying the evolutionary theories of senescence and niche conservatism. Evol. Ecol. 10 , 1–11 (1996).
Sgrò, C. M. & Partridge, L. A delayed wave of death from reproduction in Drosophila. Science 286, 2521–2524 (1999).
Lenski, R. E., Rose, M. R., Simpson, S. C. & Tadler, S. C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138 , 1315–1341 (1991).
Lenski, R. E. & Travisano, M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl Acad. Sci. USA 91, 6808–6814 (1994).
Sniegowski, P. D., Gerrish, P. J. & Lenski, R. E. Evolution of high mutation rates in experimental populations of Escherichia coli. Nature 387, 703–705 (1997).
Cooper, V. S. Consequences of ecological specialization in experimental long-term evolving populations of Escherichia coli. Thesis, Michigan State Univ. (2000).
Kimura, M. The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, 1983).
Miller, R. G. Simultaneous Statistical Inference (McGraw Hill, New York, 1981).
Cooper, V. S., Schneider, D., Blot, M. & Lenski, R. E. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of E. coli B. J. Bacteriol. (submitted).
Funchain, P. et al. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics 154, 959– 970 (2000).
De Visser, J. A. G. M., Zeyl, C. W., Gerrish, P. J., Blanchard, J. L. & Lenski, R. E. Diminishing returns from mutation supply rate in asexual populations. Science 283, 404–406 (1999).
Szathmáry, E. Do deleterious mutations act synergistically? Metabolic control theory provides a partial answer. Genetics 133, 127– 132 (1993).
Muller, H. J. The relation of recombination to mutational advantage. Mutat. Res. 1, 2–9 (1964 ).
Kondrashov, A. S. Deleterious mutations and the evolution of sexual reproduction. Nature 336, 435–440 ( 1988).
Houle, D., Hoffmaster, D. K., Assimacopoulos, S. & Charlesworth, B. The genomic mutation rate for fitness in Drosophila. Nature 359, 58–60 ( 1992).
Kibota, T. T. & Lynch, M. Estimate of the genomic mutation rate deleterious to overall fitness in E. coli. Nature 381, 694–696 (1996).
Biolog ES Microplate Instructions for Use (Biolog, Hayward, California, 1993).
Guckert, J. B. et al. Community analysis by Biolog: curve integration for statistical analysis of activated sludge microbial habitats. J. Microb. Meth. 27, 183–197 ( 1996).
Acknowledgements
We thank L. Ekunwe for assistance; J. Conner, J. Cooper, N. Cooper, D. Futuyma, D. Hall, A. Jarosz, T. Marsh, P. Moore, S. Remold and D. Rozen for discussions; and M. Blot, D. Schneider, P. Sniegowski and V. Souza for sharing unpublished data. This research was supported by NSF grants to V.S.C. and R.E.L. and by the Center for Microbial Ecology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cooper, V., Lenski, R. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407, 736–739 (2000). https://doi.org/10.1038/35037572
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35037572
This article is cited by
-
DNA uptake from a laboratory environment drives unexpected adaptation of a thermophile to a minor medium component
ISME Communications (2023)
-
Revisiting the Design of the Long-Term Evolution Experiment with Escherichia coli
Journal of Molecular Evolution (2023)
-
Evolution of Pseudomonas aeruginosa toward higher fitness under standard laboratory conditions
The ISME Journal (2021)
-
Rapid evolution destabilizes species interactions in a fluctuating environment
The ISME Journal (2021)
-
The population genetics of pathogenic Escherichia coli
Nature Reviews Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.