Abstract
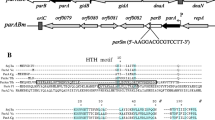
Stable inheritance of bacterial chromosomes and low-copy-number plasmids depends on the active partition of replicated molecules between daughter cells. The partition mechanism is well known for circular plasmids F and P1. The mechanism of partition of linear replicons was studied with the example of bacterio-phage N15, which persists as a linear plasmid with covalently closed ends in the lysogenic state, rather than integrating into the Escherichia coli chromosome. Since stable inheritance of N15 is due to the sop operon homologous to sop of the F plasmid, the control of expression of the N15 sop genes was analyzed. The sop promoter (Psop) contains a binding site for bacterial IHF and five CTTTGC copies, which overlap the –35 and –10 elements. The Sop proteins were shown to interact with a Psop-containing DNA fragment in vitro. Transcription of the sop operon is regulated by the Sop proteins: SopA represses Psop, and SopB enhances the repression, having no effect on the promoter activity in the absence of SopA. In N15 lysogenic cells, Psop proved to be repressed. This regulatory mechanism was assumed to ensure production of SopA and SopB in amounts required for the segregation stability of N15 and to neutralize occasional fluctuations of their concentration in the cell.
Similar content being viewed by others
REFERENCES
Austin S., Abeles A. 1983. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J. Mol. Biol. 169, 373–387.
Ogura T., Hiraga S. 1983. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 32, 351–360.
Gerdes K., Moller-Jensen J., Bugge-Jensen R. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37, 455–466.
Mori H., Mori Y., Ichinose C., Niki H., Ogura T., Kato A., Hiraga S. 1989. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J. Biol. Chem. 264, 15535–15541.
Friedman S.A., Austin S.J. 1988. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 19, 103–112.
Hayakawa Y., Murotsu T., Matsubara K. 1985. Mini-F protein that binds to a unique region for partition of mini-F plasmid DNA. J. Bacteriol. 163, 349–354.
Davis M.A., Austin S.J. 1988. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 7, 1881–1888.
Yates P., Lane D., Biek D.P. 1999. The F plasmid centromere, sopC, is required for full repression of the sopAB operon. J. Mol. Biol. 290, 627–638.
Hao J.J., Yarmolinsky M. 2002. Effects of the P1 plasmid centromere on expression of P1 partition genes. J. Bacteriol. 184, 4857–4867.
Gordon G.S., Sitnikov D., Webb C.D., Teleman A., Straight A., Losick R., Murray A.W., Wright A. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 90, 1113–1121.
Niki H., Hiraga S. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell. 90, 951–957.
Ravin V.K., Shulga M.G. 1970. Evidence for extrachromosomal location of prophage N15. Virology. 40, 800–807.
Svarchevsky A.N., Rybchin V.N. 1984. Physical DNA mapping of plasmid N15. Mol. Genet., Mikrobiol. Virusol. 10, 16–22.
Ravin N.V. 2003. Mechanisms of replication and telomere resolution of the linear plasmid prophage N15. FEMS Microbiol. Lett. 221, 1–6.
Barbour A.G., Garon C.F. 1987. Linear plasmids of the Borrelia burgdorferi have covalently closed ends. Science. 237, 409–411.
Hertwig S., Klein I., Lurz R., Lanka E., Appel B. 2003. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 48, 989–1003.
Allardet-Servent A., Michaux-Charachon S., Jumas-Bilak E., Karayan L., Ramuz M. 1993. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J. Bacteriol. 175, 7869–7874.
Malinin A.Yu., Vostrov A.A., Rybchin V.N., Svarchevsky A.N. 1992. Structure of the ends of linear plasmid N15. Mol. Genet. Mikrobiol. Virusol. 5–6, 19–22.
Ravin N., Lane D. 1999. Partition of the linear plasmid N15: interactions of N15 partition functions with the sop locus of the F plasmid. J. Bacteriol. 181, 6898–6906.
Grigoriev P.S., Lobocka M.B. 2001. Determinants of segregational stability of the linear plasmid-prophage N15 of Escherichia coli. Mol. Microbiol. 42, 355–368.
Casadaban M.J., Cohen S.N. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138, 179–207.
Grant S.G., Jessee J., Bloom F.R., Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA. 87, 4645–4649.
Ravin V., Ravin N., Casjens S., Ford M., Hatfull G., Hendrix R. 2000. Genomic sequence and analysis of the atypical bacteriophage N15. J. Mol. Biol. 299, 53–73.
Ravin N.V., Rech J., Lane D. 2003. Mapping of functional domains in F plasmid partition proteins reveals a bipartite SopB-recognition domain in SopA. J. Mol. Biol. 329, 875–889.
Simons R.W., Houman F., Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 53, 85–96.
Miller J.H. 1972. Experiments in molecular genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Lab. Press.
Sambrook J., Fritsch E.F., Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Lab. Press.
Goosen N., van de Putte P. 1995. The regulation of transcription initiation by integration host factor. Mol. Microbiol. 16, 1–7.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ravin, N.V., Dorokhov, B.D. & Lane, D. Structural Organization and Control of Expression of the sop Operon of Linear Plasmid Prophage N15. Molecular Biology 38, 247–252 (2004). https://doi.org/10.1023/B:MBIL.0000023741.96694.51
Issue Date:
DOI: https://doi.org/10.1023/B:MBIL.0000023741.96694.51