Abstract
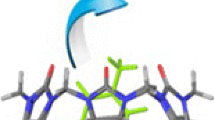
Subtilosin A produced by Bacillus subtilis is a macrocyclic peptide antibiotic which comprises 35 amino acids. Its molecular mass (3399.7 Da), determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, and chemical properties gave experimental support for unusual intramolecular linkages. The three-dimensional fold of native subtilosin in dimethylsulfoxide was determined from two-dimensional 1H-NMR spectra recorded at 600 MHz. Based on the backbone conformation, a structure for subtilosin A is presented which is characterized by three inter-residue bridges where two cysteines are linked with two phenylalanine residues, respectively, and a third cysteine is bound to a threonine residue.
Similar content being viewed by others
REFERENCES
Allen, F. H. and Kennard, O. (1993). Chem.Des.Autom.News 8, 31–37.
Arima, K., Kakinuma, A., and Tamura, G. (1968). Biochem.Biophys. Res.Commun. 31, 488–94.
Babasaki, K., Takao, T., Shimonishi, Y., and Kurahashi, K. (1985). J.Biochem. (Tokyo) 98, 585–603.
Banerjee, S. and Hansen, J. N. (1988). J.Biol.Chem. 263, 9508–9514.
Bax, A. and Davis, D. G. (1985). J.Magn.Reson. 65, 355–360.
Bax, A. and Summers, M. F. (1986). J.Am.Chem.Soc. 108, 2093–2094.
Brünger, A. T. (1996). X-PLOR version 3.851.Yale University Press, New Haven, Connecticut.
Claiborne, A., Yeh, J. I., Mallet, T. C., Luba, J., Crane, E. J., III, Charrier, V., and Parsonage, D. (1999). Biochemistry 38, 15407–15416.
Duitman, E. H., Hamoen, L. W., Rembold, M., Venema, G., Seitz, H., Saenger, W., Bernhard, F., Reinhardt, R., Schmidt, M., Ullrich, C., Stein, T., Leenders, F., and Vater, J. (1999). Proc.Natl.Acad. Sci.USA 96, 13294–13299.
Dulik, D. M. and Fenselau, C. (1987). Drug Metab.Dispos. 15, 195–199.
Gross, E., Kiltz, H. H., and Nebelin, E. (1973). Hoppe-Seyler Z.Physiol. Chem. 354, 810–812.
Hancock, R. E. and Lehrer, R. (1998). Trends Biotechnol. 16, 82–88.
Hancock, R. E. and Scott, M. G. (2000). Proc.Natl.Acad.Sci.USA 97, 8856–8861.
Jack, R. W. and Jung, G. (2000). Curr.Opin.Chem.Biol. 4, 310–317.
Jeener, J., Meier, B. H., Bachmann, P., and Ernst, R. R. (1979). J.Chem.Phys. 71, 4546–4553.
Kita, Y., Shibata, N., Kawano, N., Tohjo, T., Fujimori, C., and Ohishi, H. (1994). J.Amer.Chem.Soc. 116, 5116–5121.
Kleinkauf, H. and von Döhren, H. (1996). Eur.J.Biochem. 236, 335–351.
Koradi, R., Billeter, M., and Wüthrich, K. (1996). J.Mol.Graph. 14, 51–55.
Kunst, F., Ogasawara, N., Moszer, I., et al. (1997). Nature 390, 249–256.
Leenders, F., Stein, T. H., Kablitz, B., Franke, P., and Vater, J. (1999). Rapid Commun.Mass Spectrom. 13, 943–949.
Meyer, H. E., Heber, M., Eisermann, B., Korte, H., Metzger, J. W., and Jung, G. (1994). Anal.Biochem. 223, 185–190.
Paik, S. H., Chakicheria, A., and Hansen, J. N. (1998). J.Biol.Chem. 273, 23134–23142.
Peypoux, F., Michel, G., and Delcambe, L. (1976). Eur.J.Biochem. 63, 391–398.
Rance, M., Sørensen, O. W., Bodenhausen, G., Wagner, G., Ernst, R. R., and Wüthrich, K. (1983). Biochem.Biophys.Res.Commun. 117, 479–485.
Sahl, H. G. and Bierbaum, G. (1998). Annu.Rev.Microbiol. 52, 41–79.
Stein, T. and Vater, J. (1996). J.Biol.Chem. 271, 15428–15435.
Steller, S., Vollenbroich, D., Leenders, F., Stein, T., Conrad, B., Hofemeister, J., Jacques, P., Thonart, P., and Vater, J. (1999). Chem. Biol. 6, 31–41.
Theodoropoulos, D., Poulos, C., Gatos, D., Cordopatis, P., Escher, E., Mizrahi, J., Regoli, D., Dalietos, D., Furst, A., and Lee, T. D. (1985). J.Med.Chem. 28, 1536–1539.
Umezawa, H., Aoyagi, T., Nishikiori, T., Okuyama, A., Yamagishi, Y., Hamada, M., and Takeuchi, T. (1986). J.Antibiot. (Tokyo) 39, 737–744.
Vater, J., Stein, T., Vollenbroich, D., Kruft, V., Wittmann-Liebold, B., Franke, P., Liu, L., and Zuber, P. (1997). J.Protein Chem. 16, 557–564.
Vanittanakom, N., Loeffler, W., Koch, U., and Jung, G. (1986). J.Antibiot. (Tokyo). 39, 888–901.
Wüthrich, K. (1986). In NMR of proteins and nucleic acids. Wiley, New York, pp. 16.
Zheng, G., Hehn, R., and Zuber, P. (2000). J.Bacteriol. 182, 3266–3273.
Zimmermann, N., Metzger, J. W., and Jung, G. (1995). Eur.J.Biochem. 228, 786–797.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Marx, R., Stein, T., Entian, KD. et al. Structure of the Bacillus subtilis Peptide Antibiotic Subtilosin A Determined by 1H-NMR and Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. J Protein Chem 20, 501–506 (2001). https://doi.org/10.1023/A:1012562631268
Published:
Issue Date:
DOI: https://doi.org/10.1023/A:1012562631268