Abstract
As one of the most important food and feed crops worldwide, maize suffers much more tremendous damages under heat stress compared to other plants, which seriously inhibits plant growth and reduces productivity. To mitigate the heat-induced damages and adapt to high temperature environment, plants have evolved a series of molecular mechanisms to sense, respond and adapt high temperatures and heat stress. In this review, we summarized recent advances in molecular regulations underlying high temperature sensing, heat stress response and memory in maize, especially focusing on several important pathways and signals in high temperature sensing, and the complex transcriptional regulation of ZmHSFs (Heat Shock Factors) in heat stress response. In addition, we highlighted interactions between ZmHSFs and several epigenetic regulation factors in coordinately regulating heat stress response and memory. Finally, we laid out strategies to systematically elucidate the regulatory network of maize heat stress response, and discussed approaches for breeding future heat-tolerance maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a consequence of global warming, high temperature (HT) weather occurred more frequently than ever before with 2022 being the hottest year in record, which resulted in widespread threats to food security and agricultural sustainability (Challinor et al. 2014; Kan et al. 2023). HT could affect plant growth, development, geographical distribution, as well as crop quality and productivity (Casal and Balasubramanian 2019). In Arabidopsis, mild and moderate ambient temperatures such as 24-30 ◦C could accelerate shoot and root growth as well as the transition to flowering and other morphological changes, collectively referred to as thermomorphogenesis (Chen et al. 2022) . However, when on exposure to HT above a threshold level such as 30-37 ◦C, Arabidopsis plants typically experience heat stress (HS), impairing respiration, photosynthesis, water and nutrient uptake, and fertility (Djanaguiraman et al. 2020). Indeed, compared to the growth of un-domesticated plant species such as Arabidopsis, cereal crops are highly vulnerable to HS (Challinor et al. 2014). Every 1◦C increase in global mean temperature is estimated to reduce the global yield of wheat by 6.0%, rice by 3.2%, maize by 7.4%, and soybean by 3.1% (Zhao et al. 2017; Kraus et al. 2022). As a consequence of significant crop yield losses, it is imperative to develop crops that are capable of adaption and tolerance to HT.
As one of the most important food and feed crops worldwide, maize suffers tremendous damages under HS stress at different growth stages (Djalovic et al. 2024). At seeding stage, temperature over 30 ◦C will lead to disrupted water relation, restricted root growth and significant decrease photosynthesis rate in maize (El-Sappah et al. 2022). In reproductive stage, temperature over 35 ◦C shortens the reproductive period, decreases pollen fertility and silk elongation, thereby reducing the number of flowers and fruits, and ultimately maize yields (Sánchez et al. 2014). During pollination and grain set, temperatures over 35◦C suppress fertilization in maize and decreases its yield by 101 kg/ha per day (Dawood et al. 2020). In China's Huang-Huai-Hai and other dominant maize producing areas, HS have become a frequent natural disaster affecting the safety of maize production (Lobell et al. 2011; Hu et al. 2023). Clearly, it is of major significance to dissect the molecular basis of HS responses to identify candidate genes for heat-tolerant breeding in maize.
Although the molecular mechanisms of plant response to HT have been progressively established and reviewed in model plant Arabidopsis (Ohama et al. 2017; Ding et al. 2020; Chen et al. 2022; Guihur et al. 2022), recent studies have indicated conservative and differential regulations of HS response in major crop plants such as rice (Li et al. 2023a), wheat (Sun et al. 2021) and maize (El-Sappah et al. 2022; Djalovic et al. 2024). In this review, we summarized advances in molecular regulations underlying HT sensing and HS response in maize as well as other cereals where relevant, focusing on the complex transcriptional regulation of ZmHSFs in heat stress response. In addition, we highlighted interactions between ZmHSFs and several epigenetic regulation factors in heat stress response and memory. Finally, we laid out strategies to systematically dissect the regulatory network of maize HS response, and discussed effective approaches for future heat-tolerance maize breeding.
Pathways and signals in high temperature sensing
Pectin in the cell wall
Plants have evolved a series of molecular pathways in HT sensing and heat-induced signal cascades (El-Sappah et al. 2022). Multiple cellular and subcellular components can sense HT and subsequently activate an arrow of signaling cascades for rapid adaptive modification (Fig. 1a). For example, the cell wall is the first protective barrier in plant cells, and senses HT via a complex signal transduction (Wu et al. 2018; Wolf 2022). Moderate HS could activate PME (pectin methyl-esterase) enzyme activity in de-esterification of pectin which caused cell wall loosening and Ca2+ mobilization from apoplast to the cytoplasm (Wu et al. 2018; Wan et al. 2021). The oligogalacturonic acids derived from pectin could be further perceived by WAKs (wall-associated kinase) extracellular domain to activate MAPK (mitogen-activated protein kinases) cascade for HS sensing and response (Decreux and Messiaen 2005; Kohorn et al. 2009; Brutus et al. 2010). Recently, the expression of GhCYP703A2 and GhQRT3, two polygalacturonases were found to be inhibited under HT by affecting pectin metabolism and sporopollenin synthesis, and resulting in abnormal pollen wall development and male sterility in cotton (Li et al. 2023c). The gene expression of a wall-associated RLK-like (WAKL) gene CaWAKL20 from pepper (Capsicum annuum L.) was inhibited by HS, and silencing of CaWAKL20 could enhance pepper thermotolerance (Wang et al. 2019). Therefore, alterations of the component and modification of pectin could affect plant HT sensing by remodeling cell wall structure, and the molecular regulation of pectin synthesis and other related genes such as ZmWAKs need to be further studied in maize.
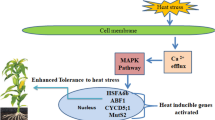
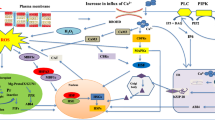
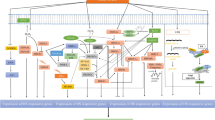
The complex molecular regulations of heat stress response in plants. a The major pathways and signals in high temperature sensing including pectin from cell wall, Ca2+ signaling channels and sensors in plasma membrane, reactive oxygen species and other nuclear proteins. b The transcriptional regulation of heat stress response mainly controlled by HSF-HSP pathway in maize. Several ZmHSFs have been identified positively in regulating HS response in plants including ZmHSF01, ZmHSF04, ZmHSF05, ZmHSF06 and ZmHSF17, while ZmHSF08, ZmHSF11and ZmHSF25 negatively regulate HS response. Other proteins such as ZmDREB2A and ZmbZIP60 also play functions in heat stress response. Several HSFs could coordinately regulate HS response through protein interaction such as HSFA2 and HSFA3. c The epigenetic regulations in heat stress memory. Several DNA methyltransferases such as CMT2, is considered an inhibitory mark in gene expression silencing of HSFs and HSPs. Histone H3 methylated on K4 (H3K4me) and H3K36me mainly generates an open chromatin configuration related to transcription activation. Several genes such as the histone acetyltransferase GCN5 (general control non-depressible 5) promote open chromatins and enhance gene expression. H3K9me3 and H3K27me3 are responsible for closed chromatin states that result in transcriptional repression. Several genes such as JUMONJI (JMJ) proteins, HISTONE DEACETYLASE9 (HDA9) could maintain repressive histone marks on memory genes and repress gene expression of HSFs. Several non-coding RNAs trans-acting small interfering RNAs (ta-siRNAs), microRNAs (miRNAs), and long non-coding circular RNAs (lncRNAs) also play an indispensable role in regulating heat stress response
Thermosensors at plasma membrane
Increasing evidence proves the primary HT sensing occurs at the plasma membrane through many thermosensors in plants (Bourgine and Guihur 2021; Mittler et al. 2011). Any protein could be defined as a thermosensor not only able to perceive high temperature increments but also trigger specific signaling pathways which can upregulate related heat stress response (HSR) genes (Vu et al. 2019; Mittler et al. 2011). For example, Cyclic Nucleotide-Gated Channels (CNGCs) act as thermosensors embedded in the plasma membrane to allow the inward flux of calcium and activate the HS response (Finka et al. 2012). There are 11 plasma membrane-localized cyclic nucleotide-gated ion channels (CNGC) genes identified to be Ca2+ conducting channels of maize in heat sensing (Hao and Qiao 2018). In response to heat, Ca2+ acts as an important messenger that binds directly to CaM (Calmodulin) and change their conformation to elicit HSR (Dodd et al. 2010). CaM-binding protein kinase CBK3 is a positive regulator in the HS signal transduction by phosphorylating HsfA1 and enhancing its DNA-binding activity (Liu et al. 2018b). ZmCDPK7 is another plasma membrane-anchored protein under normal conditions, but could be also localized to the cytoplasm under HS to induce Reactive Oxygen Species (ROS) accumulation and enhance heat-stress tolerance by phosphorylating sHSP17.4 and respiratory burst oxidase homolog RBOHB in maize (Zhao et al. 2021). Recently, a new sensor from plasma membrane, Thermo-tolerance 3 (TT3) , consisting of two genes, plasma membrane–localized E3 ligase TT3.1 and chloroplast precursor protein TT3.2, could interact together in endosome to promote the degradation of TT3.2 to protect chloroplast from heat stress damage and reduce grain-yield losses under heat stress in rice (Zhang et al. 2022; Li and Liu 2022a). In future studies, it would be exciting to identify more sensors of HS located in plasma membrane in major crop plants, such as maize.
Reactive oxygen species as signal molecules
ROS is an important signaling molecule that could stimulate or interact with the Ca2+ signaling pathways during heat stress sensing (Li et al. 2018). HS alters the normal status of the chloroplast and mitochondria membranes, causing the over-accumulation of ROS (Navarro et al. 2021). For example, NDL1 encodes a mitochondria localized ATP-dependent metalloprotease essential for regulating thermotolerant maize growth by altering endogenous auxin levels, and needle1 (ndl1) is a temperature-sensitive mutant with hyper-accumulate ROS owing to respiratory defects in mitochondria (Liu et al. 2019a). When the accumulated ROS elicit cell damage, antioxidants and ROS scavenging genes are activated to maintain cellular redox homeostasis and HS acclimation (Choudhury et al. 2017). In response to ROS, HSR genes, such as several MYBs, AP2/EREBPs, NACs, HSPs, Rubisco and APX and are activated to scavenge the accumulated ROS (Jagtap et al. 2020; Khan et al. 2023). ROS may activate HSFA1 by inducing their trimerization, which, in turn, enhanced their DNA binding activity with the targeting HSR genes (Liu et al. 2013). Therefore, it is important to explore and identify the genes which generate or eliminate ROS for HT sensing and responsing in maize.
Other proteins
Other proteins are also participated in thermosensing. Phytochromes function in thermosensing and signal transduction pathways during heat stress (Jung et al. 2016). PhyB (Phytochrome B) signaling is exacerbated by warm temperature during early night in Arabidopsis (Vu et al. 2019). The basic-helix-loop-helix transcription factors PIF4 (PHYTOCHROME-INTERACTING FACTOR 4) and PIF7 are two master regulators in thermomorphogenesis (Koini et al. 2009; Kumar et al. 2012). PIF4, is involved in promoting auxin-dependent growth in warm conditions (Zhao and Bao 2021). The protein level of PIF7 is enhanced by conformational changes in its RNA secondary structures in response to higher temperatures in Arabidopsis (Chung et al. 2020). ELF3 (EARLY FLOWERING 3) containing a prion-like domain functions as a repressor and negative regulator of thermomorphogenesis in Arabidopsis (Jung et al. 2020). Moreover, several hormones have multiple functions in the response to HS. EIN3 (ETHYLENE-INSENSITIVE 3) is a key transcriptional regulator of the ethylene signaling pathway and higher temperatures promote EIN3 protein accumulation (Hao et al. 2021). BRASSINAZOLE RESISTANT1 (BZR1) is a positive regulator of BR (Brassinosteroid) signaling, and over-expression of BZR1, improves thermotolerance by enhancing H2O2 levels in tomato (Yin et al. 2018). In maize, these related proteins are less studied to regulate HT sensing. In the future, more genes related to other traits of HS sensing could be identified in different maize populations by quantitative trait locus (QTLs) mapping and genome-wide association study (GWAS). For example, four QTLs and 17 genes associated with 42 single nucleotide polymorphisms (SNPs) related to thermotolerance of seed-set had been identified by mapping of QTLs and GWAS of 261 diverse maize lines (Gao et al. 2019).
The transcription regulation of HS response
HSFs act as master transcription factors in HS response
After plants sense HS signals, a primary response occurs mainly by heat-induced proteins such as heat shock transcription factors (HSFs) to active downstream heat shock proteins (HSPs) and other HSR gene expression to mitigate the effect of HS, which is conserved across different species (El-Sappah et al. 2022; Guihur et al. 2022). Plants have evolved multiple HSFs to participate in HS response, which are divided into three classes: HSFA, HSFB, and HSFC according to their structural characteristics and phylogenetic comparisons (Lin et al. 2011; Zhang et al. 2020a). Class A HSFs contribute to transcriptional activation, whereas the rest two classes have no specific role in transcriptional activation and might be serving as coactivators cooperating with class A (Haider et al. 2022). HSFA1 is the master transcription activator inducing expression of different HSFs by binding the HS elements in their promoter regions, including dehydration responsive element binding protein2A (DREB2A), ERF/AP2, HSFA2, HSFA7 and HSFBs, which has been proved to be conservative in different plant species (Yoshida et al. 2011). HSFA1 activates HSFA2 and prolongs the acquired thermotolerance by maintaining HSP expression for HS generational memory (acquired thermotolerance for several days) in Arabidopsis (Charng et al. 2023). HSFA1 could interact with HSP70 and HSP90 to repress HSFA1 activity under normal conditions in tomato (Hahn et al. 2011). HSFA1 proteins could also regulate self-activity by post-translational modification, phosphorylation and SUMOylation in Arabidopsis and wheat (Rytz et al. 2018; Ding et al. 2020; Wang et al. 2023). HSFA1s are required for PIF4 accumulation at a warm daytime temperature through protein interaction, and play a critical regulator in integrating both thermomorphogenesis and HS responses in Arabidopsis (Li et al. 2024a; Tan et al. 2023 Science Advance). HSFA1 was also regulated by miR165 and miR166 and their target transcript, PHABULOSA (PHB), at the transcriptional and translational levels to control plant HS responses in Arabidopsis (Li et al. 2023b). What is more, the HS-induced epigenetic states, can be transmitted to the next generation (transgenerational memory which is another type of HS memory) through meiosis (Liu et al. 2019b; Zhu et al. 2023). Therefore, the HSF gene family plays an important and complex transcription regulation on downstream HSPs and HSR gene expression in HS response and memory.
Identification and function study of ZmHSFs
In maize, there are 31non-redundant HSFs and might have multiple regulations on different stress responses (Zhang et al. 2020a). We summarized recent functional studies of ZmHSFs as shown in Fig. 1b and Table 1. ZmHSF01, ZmHSF03, and ZmHSF23 was observed with higher expression exposed to heat stress probably proving their significant roles in regulating heat stress response (Lin et al. 2011). In Arabidopsis seedlings, ZmHSF01 (ZmHSFTF13) compensated for the thermotolerance defects of mutant athsfa2, and ZmHSF01-overexpressing lines showed enhanced basal and acquired thermotolerance (Zhang et al. 2020b). ZmHSFA2 (ZmHSF04) and HEAT SHOCK BINDING PROTEIN 2 (ZmHSBP2) physically interact with each other and antagonistically modulate expression of GALACTINOL SYNTHASE2 (ZmGOLS2) and raffinose biosynthesis in transformed maize protoplasts and Arabidopsis plants (Gu et al. 2019). Ectopic overexpression of ZmHSF04 confers increased thermal and salt‑stress tolerance in transgenic Arabidopsis by up-regulated the expression of heat-specific HSP genes (AtHsp25.3, AtHsp18.2, and AtHsp70B) and stress-related genes (AtAPX2 and AtGolS1) compared to the wild type (Jiang et al. 2017). ZmHSF05 improves thermotolerance by up-regulating Hsps in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant (Li et al. 2019b). ZmHSF06 is the specific gene localized in pollens and regulate Hsp70-2 and Hsp70-4 in response to HS (Li et al. 2015). Overexpression of ZmHSF06 in Arabidopsis plants have enhanced basal and acquired thermotolerance, stronger drought-stress tolerance and growth advantages under mild heat stress conditions (Li et al. 2015). Overexpressing of ZmHSF17-II, an intron retention isoform of subclass A2 gene ZmHSF17, could increase the thermotolerance in Arabidopsis (Zhang et al. 2024). ZmHSF08 from class B HSFs was induced by salt, drought, and abscisic acid (ABA) treatment, and negatively regulates many stress/ABA response genes under salt stress and drought stress (Wang et al. 2021). ZmHSF11, also a member of class B HSFs, decreases plant tolerance to heat stress by negatively regulating the expression of oxidative stress-related genes HSP17 and HSFA2e (Qin et al. 2022). Recently, ZmHSF20 of class B2a was reported as a negative factor in regulating heat stress through the ZmHSF20-ZmHSF4-ZmCESA2 module (Li et al. 2024b). ZmHSF20 could bind the promoters of ZmCESA2 and ZmHSF4, while ZmHSF4 directly activated the expression of ZmCESA2 to positively regulate heat response (Li et al. 2024b). Although many ZmHSFs have been cloned and studied in Arabidopsis, the real regulatory functions of ZmHSFs in response to different stress need to be validated in maize by creating overexpressing and knockout transgenic plants in maize. Furthermore, the down-regulated and interaction network of ZmHSFs in responses to HS need to be systematically studied to extend our understanding of the complexity of the HS regulation in maize beyond Arabidopsis.
The interactions between HSFs in coordinately regulating heat stress response
There is increasing evidence for the coordinate regulation of HSFs in HS response by protein interaction. The interactions could exist in different isoforms of one HSF, or the same subfamily of HSFs or different subfamily of HSFs. For example, ZmHSF17-I and ZmHSF17-II, two different isoforms of ZmHSF17 by alternative splicing, could interacted with each other to negatively regulate its own transcription under heat stress (Zhang et al. 2024). HSFA3 could interact with the same sub-family gene HSFA2 to form heteromeric complexes that efficiently promotes transcriptional memory genes such as HSA32, APX2 and HSP22 by influencing histone H3 lysine4 (H3K4) hyper-methylation following HS exposure in Arabidopsis (Friedrich et al. 2021). Recently, LIHSF2C, a class C HSF involved in thermotolerance, could not only interact with itself, but also interact with LlHSFAs of lily, AtHSFA1e and AtHSFA2 of Arabidopsis, and NbHSFA2 of tobacco (Wu et al. 2024). After suffering HS, the homologous interaction of LlHSFC2 was repressed, but the heterologous interaction between LlHSFC2 and HSFAs was promoted, which exerted its co-activation effect for thermotolerance establishment and maintenance (Wu et al. 2024). Even though theses interactions between HSFs have been identified in many plants, the coordinated mechanisms need to be further explored to understand the fine regulation of HS response especially in maize.
HSPs, not just downstream genes of HSFs in HS response
Heat shock proteins (HSPs) have multiple functions in HS response and regulation. First, HSPs including HSP20, HSP60, HSP70, HSP90, HSP100, and HSP110 are induced by HSFs, and act as chaperones to renature misfolded proteins that are important for thermotolerance in Arabidopsis and other plants (Lee et al. 2007; Kotak et al. 2007; EI-Sappah et al. 2022). However, single mutants of these HSPs do not cause severe HS-deficient phenotype indicating the functional redundance of HSPs in HS response (Guihur et al. 2022). HSP101 is one of key HSPs for the acquisition of thermotolerance in different plants (Gurley 2000). In rice, OsHSP101 acts as positive regulator of thermotolerance and heat memory in rice (Lin et al. 2014). Transcriptome data showed that ZmHSP101 is highly expressed in male meiocytes under normal growth conditions (Dukowic-Schulze and Chen 2014). Recently, ZmHSP101 was found to function in DNA double-strand breaks repair and subsequent meiosis. Overexpression of ZmHSP101 in anthers results in robust microspores with enhanced heat tolerance (Li et al. 2022c). In addition, abscisic acid-induced calcium-dependent protein kinase ZmCDPK7 interacts with the small heat shock protein sHSP17.4 through phosphorylation, and participates in thermotolerance in maize (Zhao et al. 2021).
In addition, multiple isoforms of HSP and HSF could form complexes and function downstream in determining transcriptional levels. For example, to prevent HSFA1 proteins from being activated under normal temperature conditions, HSP70 forms a complex with HSFA1s, repressing their DNA binding activity and nuclear localization of HSFA1 under permissive temperatures, while HS activates HSFA1 by loosening the complex in tomato (Yamada et al. 2007; Hahn et al. 2011; Ding et al. 2020). Although HSPs are crucial in the regulation of HS, the specific functions with HSFs and targets of HSPs in major crop plants are largely not yet clear.
Other transcription factors in regulating heat stress response
Beside HSF family genes, some other different transcription factors also play functions in HS response in plants. A lily membrane-associated NAC transcription factor LlNAC014 increased thermotolerance by activating the DREB2-HSFA3 module in lily (Lilium longiflorum) (Wu et al. 2023). DREB2A (DEHYDRATION-RESPONSIVE ELEMENT-BINDING2A) could regulate the expression of HSFA3 and downstream HSR genes by forming a trimer complex comprised of NUCLEAR FACTOR Y, SUBUNIT A2 (NF-YA2), DNA POLYMERASE II SUBUNIT B3-1(DPB3-1), and NF-YB3 in Arabidopsis (Schramm et al. 2008; Ohama et al. 2017; Ding et al. 2020). BRI1-EMS-SUPPRESSOR 1 (BES1), which is a key regulator of brassinosteroids (BRs) signalling, could be de-phosphorylated mediated by ABA-controlled PP2C phosphatases and activated even in the absence of BRs by heat (Yao et al. 2022). Furthermore, the activated BES1 could interact with HSFA1a and enhance its binding activity to targeted HSEs whether or not under the regulation of ABA or BR (Albertos et al. 2022). REVEILLE4 (RVE4) and REVEILLE8 (RVE8) are two transcription factors of Arabidopsis involved in regulating circadian clock and early HS-induced gene expression, indicating that plants can redeploy and coordinate multiple regulatory networks to adapt to the changing environment (Li et al. 2019a). Heat stress bzip28 and bzip60 double-mutant plants are sensitive to heat stress, and ZmbZIP60 activates the expression of a type-A HSFTF13, which, in turn, upregulates the expression of a constellation of HSP genes (Li et al. 2020b). ZmDREB2A plays an essential role both in both heat and drought tolerance in maize (Qin et al. 2007). These results indicate heat stress is connected with other abiotic stresses, and plants have evolved other compensating pathways except HSFs to adapt and combat heat stress.
The interactions between HSFs and epigenetic factors in regulating heat stress memory
Heat stress memory
HS memory is an important and active process that allow plants to acquire thermotolerance to respond more efficiently when plants encounter stresses more than once. Plants have evolved several HS memory strategies, including generational memory (over several days or weeks) (Perrella et al. 2022; Balazadeh 2022) and transgenerational memory (transmitted to the next generation) (Gallusci et al. 2023). Epigenetic regulation of the HS mainly includes DNA methylation, histone modification and modulation by small non-coding RNA. These epigenetic modifications can activate or repress transcription by generating either 'open' or 'closed' chromatin configurations to regulate the accessibility of HSF transcriptional regulators to HSR genes (Ueda and Seki 2020). We evaluated recent epigenetic modifications of HSF and HSR genes in regulating HS memory as shown in Fig. 1c and Table 2.
DNA methylation in response to HS
Methylation is the most common form of DNA modification, involving the addition of methyl groups to DNA molecules, thereby altering DNA structure and gene expression, ultimately impacting the plant growth, development, and responses to stressors (Law and Jacobsen 2010; Liu et al. 2023a). 5mC modification catalyzed by DNA methyltransferases, and is considered an inhibitory mark in gene expression, transposon insertion, and excision, as well as genome stability (Kong et al. 2023). HS could induce DNA hypomethylation and hypermethylation. On the other hand, these marked DNA may be carried to the next generation making the progenies ‘primed’ for HS responses (Arora et al. 2022). For example, natural CMT2 (Chromomethylase 2) variation is associated with genome-wide methylation changes, and cmt2 mutants shew more tolerance to heat-stress, which suggests genetic regulation of DNA epigenetic modifications as a likely mechanism underlying natural adaptation to high temperatures (Shen et al. 2014). However, it still lacks the epigenomic landscape of DNA methylation in response to HS in maize.
Histone methylation in regulating HSR genes
Histone methylation is another epigenetic mechanism that plays a key role in mediating plant responses to HS (He et al. 2021). Current researches on histone methylation mainly focus on histones H3 and H4, in which H3K4, K9, K27, and K36 are methylated (He et al. 2021). Histone H3 methylated on K4 (H3K4me) and K36 (H3K36me) mainly generates an open chromatin configuration related to transcription activation, while H3K9me and H3K27me are responsible for closed chromatin states that result in transcriptional repression (Liu et al. 2010; Ueda and Seki 2020). After a treatment of mild primary HS, di- and tri-methylated histone H3 (H3K4me2 and H3K4me3) were enriched at loci of HS memory genes such as APX2, HSP18.2 and HSP22, to provide higher and longer expression of these HS memory genes for remembering acquired thermotolerance (Liu et al. 2018a; Shekhawat et al. 2021). JUMONJI (JMJ) proteins such as JMJ30 and JMJ12, are demethylases involved in regulating H3K27me3, and could maintain repressive histone marks on HT memory genes in Arabidopsis (He et al. 2021; Song et al. 2021; Yamaguchi et al. 2021). So, it is interesting to identify genes regulating histone methylation and demethylation in response to HT memory in maize in future studies.
Histone acetylation in HS memory
Besides methylation, histone acetylation also plays an important role in plant heat response. Acetylation of histone weakens the interaction of DNA with histone and promotes chromatin decondensation to enhance the transcriptional activity. Histone deacetylation is mainly catalyzed by histone deacetylases (HDACs), which play different roles in response to HS. HD2C acts as a transcriptional repressor of heat-activated genes by removing lysine acetylation at chromatin loci of heat activated genes (Buszewicz et al. 2016). HDA9 (HISTONE DEACETYLASE 9) relocates from the cytosol to the nucleus and positively modulates heat shock signal transduction (Niu et al. 2022). It is also proved that this mechanism of heat-induced cytoplasm-to-nucleus translocation of HDA9 is conserved in wheat and rice, indicating its potential use in crop breeding during global climate warming (Niu et al. 2022). In addition, HDA9 keeps stabilized in response to HT and mediates the histone deacetylation of YUCCA8, an important enzyme in auxin biosynthesis, and promotes the eviction of H2A.Z from nucleosomes resulting in the binding and transcriptional activation of PHYTOCHROME INTERACTING FACTOR 4, revealing the crosstalk between histone modification and chromatin remodeling in HT responses (van der Woude et al. 2019). The functions of these HAD genes in maize remain unknown and need to be further studied in regulating HT response and memory.
Non-coding RNAs in HS memory
Non-coding RNAs including trans-acting small interfering RNAs (ta-siRNAs), microRNAs (miRNAs), and long non-coding circular RNAs (lncRNAs) play an indispensable role in regulating HS (Liu et al. 2015). Several miRNAs such as miR398, miR156, miR159 and miR160 are found to be induced by HS in Arabidopsis, poplar and wheat (Sunkar et al. 2012). Under HS, the abundance of ta-siRNAs-trans-acting siRNA precursor 1 (TAS1) was decreased leading to higher expression of HTT gene expression levels which is cofactors of HEAT SHOCK PROTEIN 70-14 (HSP70-14) to mediate thermotolerance (Li et al. 2014). The OsSGS3-ta-siRNA-OsARF3 module orchestrates trade-offs between thermotolerance and defense in rice (Gu et al. 2023). Other microRNAs such as miR398 and miR156 are positive regulators of some important transcription factors and downstream genes such as CSDs (Copper/zinc Superoxide Dismutase) and SPL13 (Squamosa Promoter-Binding Protein-Like 13) to maintain thermotolerance (Matthews et al. 2019; Li et al. 2020a). When plants suffer HT damage, lncRNAs can respond to stress through various mechanisms, including the accumulation of osmoprotectants (e.g., proline), calcium ion regulation, stomatal regulation, and hormone signaling and synthesis (Song et al. 2020). Recently, another category of small RNAs called circRNA was found to be involved in HS possibly by interacting with other regulators to control the expression of the HSR genes (He et al. 2020). In maize, by applying a high-throughput sequencing of IncRNAs and sRNA in inbred line CM1, 993 lncRNAs and 340 miRNAs were identified with significantly differential expression under heat stress compared to normal conditions, and constructed a lncRNA-mediated regulatory network to help visualize the molecular response mechanism of IncRNA and mi-RNA in response to heat stress (Zhao et al. 2022; Hu et al. 2022).
The coordination between HSFs and epigenetic factors in regulating HS memory
Plants have evolved a complex molecular mechanism in retaining, sustaining and transmitting HS memories, especially by the coordination between HSFs and epigenetic factors (Sharma et al. 2022; Zhu et al. 2023). For example, HSFA2 plays a key role in regulating HS memory by cooperating with histone methylation. HSFA2 is necessary for the sustained accumulation of H3K4 hypermethylation at the promoter region of HS memory genes in response to repeated heat stress (Lämke et al. 2016). This mechanism is only transiently associated with heat stress memory loci in the hours following heat stress. In addition, HSFA2 could also activate H3K27me3 demethylase RELATIVE OF EARLY FLOWERING 6 (REF6), and feedback form a positive loop to transmit transgenerational memory of heat by derepressing HSFA2 (Liu et al. 2019b). Another example is the histone acetyltransferase GCN5 (GENERAL CONTROL NON-DEPRESSIBLE 5) enhanced levels of histone H3 acetylation (H3K9/14ac) at the promoter regions of the HSFA3 and UV-HYPERSENSITIVE 6 (UVH6) genes leading to higher transcription of HSFA2, HSFA3, and UVH6 and confers thermotolerance in Arabidopsis (Hu et al. 2015). What is more, siRNAs are also mediate the silencing of a Copia-type retrotransposon named ONSEN which could be bound by HSFA2 through heat-response element (Cavrak et al. 2014). Therefore, HSF-mediated epigenetic regulation may be a widely adopted mechanism in heat stress memory in plants, which need to be studied in maize.
Conclusions and perspective
The complex transcriptional regulation network of heat stress response in maize
Since high temperature and heat damage significantly affect plant growth, development, crop yield and quality, especially in maize, it is imperative to understand the molecular mechanisms and regulatory networks involved in the HS response. Plants have evolved a series of molecular mechanisms to sense heat stress accurately and induce different genes and signal cascades (El-Sappah et al. 2022). The transcriptional regulation by heat-induced HSFs is as a primary heat response to activate HSPs and other HSR genes to mitigate the effect of heat stress. Epigenetic regulation is also pivotal for heat stress responses, especially in heat stress memory. Furthermore, more studies revealed the coordination between different transcription factors and epigenetic factors is essential for the precise regulation for plant temperature responses and phenological adaption (Zhu et al. 2023). Although these mechanisms have been well studied in Arabidopsis, the conservative and differential regulation in other crop plants such as maize need to further explained. In the future, more transcriptional and epigenetic factors related to HS could be further identified by constructing the gene regulatory network and epigenetic landscape of HS, and their complex interactions of HS response need to be systematically explained to decipher the complex transcriptional regulation network of heat stress response in maize.
Multi-omics help dissect the complex regulatory network in heat stress response
With the development of genome sequencing, multi-omics data helps to dissect multiple regulation pathways in different tissues under HT and heat stress (Derbyshire et al. 2022) (Fig. 2a). Compared to the regulation of leaves in response to HT, the mechanisms of pollen and silk in response to heat stress sense more important in relation to male sterility and crop yield (Chaturvedi et al. 2021; Khan et al. 2023). Single-cell multi-omics, including genomics, transcriptomics, epigenomics, and even proteomics, will provide unprecedented perspectives on the regulatory mechanisms of plant development and physiological responses (Yu et al. 2023). For example, the dynamic development atlas of maize pollen and meristem in response to HT would be interesting to be studied by using single-cell multi-omics to discover new cell types and new regulatory mechanisms acting during stress responses (Zhu et al. 2023). High throughput methods needed to be developed to profile the dynamic TFs and epigenetic binding landscape. For example, a low-cost and high-throughput in vivo chromatin profiling method tsCUT&Tag (a transient and simplified cleavage under targets and tagmentation) could have great potential for profiling the transcription factor binding landscape across plant development (Wu et al. 2021b). In addition, machine learning approaches and high-throughput phenomics coupled with precision gene-editing in crop species are likely to aid the breeding of stress-tolerant varieties to confront the daunting challenges of the incoming decades (Wu et al. 2021a). The integration of multi-omics with systems biology would be helpful to identify potential candidate genes for crop improvements under environmental stress conditions (Yang et al. 2021) (Fig. 2b).
Systematic strategies to construct the regulatory network of heat stress response and enhance heat-tolerance maize breeding. a Multi-omics to study heat stress response of maize including genomics, transcriptomics, epigenomics and metabolomics. b The construction of regulatory network of ZmHSFs based on multi-omics data. c Modern genetic approaches to identify heat-tolerance quantitative trait locus or elite haplotypes for promoting heat-tolerance maize breeding
The combination study of heat with drought stresses
As global climate change, the combined stresses of high temperature and drought occurred more frequently and extensively, which make more threats to crop growth and productivity (Hu et al. 2023). Maize is a sensitive crop to both drought and heat stresses, particularly at the reproductive stages of development. Although major progress has been made in understanding the molecular regulation of drought and heat, respectively during past decades (EI-Sappah et al. 2022; Singh et al. 2023; Yang et al. 2023), the influence and molecular regulations of the combined stresses of high temperature and drought on summer maize production remains largely unknown. Therefore, the combination study of maize in response to heat with drought stresses should be a focus for cultivating elite maize verities with maximum tolerance against drought and heat stress in the future.
Modern genetic approaches for heat-tolerant crop breeding
Modern genetic approaches, such as QTL-mapping, bulked segregant analysis (BSA), GWAS and genome editing, have facilitated gene cloning to develop maize verities with the highest heat tolerance (Benavente and Giménez 2021). The identification of heat-tolerance quantitative trait loci or natural variations by next-generation bulked segregant analysis for breeding 4.0 from wild and landrace species or other genetic population materials may provide effective ways to breed heat-tolerant crops (Wang et al. 2023) (Fig. 2c). In addition, gene editing technologies based on the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein (Cas) system have great potential to create much more variable sites and elite haplotypes for promoting heat tolerance by the manipulation of HSR genes (Zhu et al. 2023). Genetic variability and the effect of heat stress on crops in the field also need to be thoroughly investigated in order to provide useful strategies that can ensure future plants to successfully adapt to environmental temperature fluctuations caused by global climate change.
Availability of data and materials
Not applicable.
Abbreviations
- ABA:
-
Abscisic acid
- BSA:
-
Bulked segregant analysis
- ChIP-seq:
-
Chromatin immunoprecipitation sequencing
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- ER:
-
Endoplasmic Reticulum
- GWAS:
-
Genome-wide association study
- HSR:
-
Heat stress response
- HSF:
-
Heat shock factor
- HSP:
-
Heat shock protein
- HT:
-
High temperature
- lncRNA:
-
Long non-coding circular RNA
- miRNA:
-
MicroRNA
- QTL:
-
Quantitative trait locus
- ROS:
-
Reactive oxygen species
- SNP:
-
Single nucleotide polymorphism
- ta-siRNA:
-
Trans-acting small interfering RNA
References
Albertos P, Dündar G, Schenk P, Carrera S, Cavelius P, Sieberer T, Poppenberger B (2022) Transcription factor BES1 interacts with HSFA1 to promote heat stress resistance of plants. EMBO J 41(3):e108664. https://doi.org/10.15252/embj.2021108664
Arora H, Singh RK, Sharma S, Sharma N, Panchal A, Das T, Prasad A, Prasad M (2022) DNA methylation dynamics in response to abiotic and pathogen stress in plants. Plant Cell Rep 41(10):1931–1944. https://doi.org/10.1007/s00299-022-02901-x
Balazadeh S (2022) A ‘hot’ cocktail: the multiple layers of thermomemory in plants. Curr Opin Plant Biol 65:102147. https://doi.org/10.1016/j.pbi.2021.102147
Benavente E, Giménez E (2021) Modern approaches for the genetic improvement of rice, wheat and maize for abiotic constraints-related traits: a comparative overview. Agronomy 11:376. https://doi.org/10.3390/agronomy11020376
Bourgine B, Guihur A (2021) Heat shock signaling in land plants: from plasma membrane sensing to the transcription of small heat shock proteins. Front Plant Sci 12:710801. https://doi.org/10.3389/fpls.2021.710801
Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107(20):9452–9457. https://doi.org/10.1073/pnas.1000675107
Buszewicz D, Archacki R, Palusiński A, Kotliński M, Fogtman A, Iwanicka-Nowicka R, Sosnowska K, Kuciński J, Pupel P, Olędzki J, Dadlez M, Misicka A, Jerzmanowski A, Koblowska MK (2016) HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ 39(10):2108–2122. https://doi.org/10.1111/pce.12756
Casal JJ, Balasubramanian S (2019) Thermomorphogenesis. Annu Rev Plant Biol 70:321–346. https://doi.org/10.1146/annurev-arplant-050718-095919
Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten Scheid O (2014) How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet 10(1):e1004115. https://doi.org/10.1371/journal.pgen.1004115
Challinor AJ, Watson J, Lobell DB, Howden SM, Smith DR, Chhetri N (2014) A meta-analysis of crop yield under climate change and adaptation. Nat Clim Change 4:287–291. https://doi.org/10.1038/nclimate2153
Charng YY, Mitra S, Yu SJ (2023) Maintenance of abiotic stress memory in plants: Lessons learned from heat acclimation. Plant Cell 35(1):187–200. https://doi.org/10.1093/plcell/koac313
Chaturvedi P, Wiese AJ, Ghatak A, Záveská Drábková L, Weckwerth W, Honys D (2021) Heat stress response mechanisms in pollen development. New Phytol 231(2):571–585. https://doi.org/10.1111/nph.17380
Chen Z, Galli M, Gallavotti A (2022) Mechanisms of temperature-regulated growth and thermotolerance in crop species. Curr Opin Plant Biol 65:102134. https://doi.org/10.1016/j.pbi.2021.102134
Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J 90(5):856–867. https://doi.org/10.1111/tpj.13299
Chung BYW, Balcerowicz M, Di Antonio M, Jaeger KE, Geng F, Franaszek K, Marriott P, Brierley I, Firth AE, Wigge PA (2020) An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat Plants 6(5):522–532. https://doi.org/10.1038/s41477-020-0633-3
Dawood MFA, Moursi YS, Amro A, Baenziger PS, Sallam A (2020) Investigation of heat-induced changes in the grain yield and grains metabolites, with molecular insights on the candidate genes in Barley. Agronomy 10(11):1730. https://doi.org/10.3390/agronomy10111730
Decreux A, Messiaen J (2005) Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol 46:268–278. https://doi.org/10.1093/pcp/pci026
Derbyshire M, Batley J, Edwards D (2022) Use of multiple ‘omics techniques to accelerate the breeding of abiotic stress tolerant crops. Curr Plant Biol 32:100262. https://doi.org/10.1016/j.cpb.2022.100262
Ding Y, Shi Y, Yang S (2020) Molecular regulation of plant responses to environmental temperatures. Mol Plant 13(4):544–564. https://doi.org/10.1016/j.molp.2020.02.004
Djalovic I, Kundu S, Bahuguna RN, Pareek A, Raza A, Singla-Pareek SL, Prasad PVV, Varshney RK (2024) Maize and heat stress: Physiological, genetic, and molecular insights. Plant Genome 17(1):e20378. https://doi.org/10.1002/tpg2.20378
Djanaguiraman M, Narayanan S, Erdayani E, Prasad PVV (2020) Effects of high temperature stress during anthesis and grain filling periods on photosynthesis, lipids and grain yield in wheat. BMC Plant Biol 20(1):268. https://doi.org/10.1186/s12870-020-02479-0
Dodd AN, Kudla J, Sanders D (2010) The language of calcium signaling. Annu Rev Plant Biol 61:593–620. https://doi.org/10.1146/annurev-arplant-070109-104628
Dukowic-Schulze S, Chen C (2014) The meiotic transcriptome architecture of plants. Front Plant Sci 5:220. https://doi.org/10.3389/fpls.2014.00220
El-Sappah AH, Rather SA, Wani SH, Elrys AS, Bilal M, Huang Q, Dar ZA, Elashtokhy MMA, Soaud N, Koul M, Mir RR, Yan K, Li J, El-Tarabily KA, Abbas M (2022) Heat stress-mediated constraints in maize (Zea mays) production: challenges and solutions. Front Plant Sci 13:879366. https://doi.org/10.3389/fpls.2022.879366
Finka A, Cuendet AF, Maathuis FJ, Saidi Y, Goloubinoff P (2012) Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24(8):3333–48. https://doi.org/10.1105/tpc.112.095844
Friedrich T, Oberkofler V, Trindade I, Altmann S, Brzezinka K, Lämke J, Gorka M, Kappel C, Sokolowska E, Skirycz A, Graf A, Bäurle I (2021) Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat Commun 12(1):3426. https://doi.org/10.1038/s41467-021-23786-6
Gallusci P, Agius DR, Moschou PN, Dobránszki J, Kaiserli E, Martinelli F (2023) Deep inside the epigenetic memories of stressed plants. Trends Plant Sci 28(2):142–153. https://doi.org/10.1016/j.tplants.2022.09.004
Gao J, Wang S, Zhou Z, Wang S, Dong C, Mu C, Song Y, Ma P, Li C, Wang Z, He K, Han C, Chen J, Yu H, Wu J (2019) Linkage mapping and genome-wide association reveal candidate genes conferring thermotolerance of seed-set in maize. J Exp Bot 70(18):4849–4864. https://doi.org/10.1093/jxb/erz171
Gu L, Jiang T, Zhang C, Li X, Wang C, Zhang Y, Li T, Dirk LMA, Downie AB, Zhao T (2019) Maize HSFA2 and HSBP2 antagonistically modulate raffinose biosynthesis and heat tolerance in Arabidopsis. Plant J 100(1):128–142. https://doi.org/10.1111/tpj.14434
Gu X, Si F, Feng Z, Li S, Liang D, Yang P, Yang C, Yan B, Tang J, Yang Y et al (2023) The OsSGS3-tasiRNA-OsARF3 module orchestrates abiotic-biotic stress response trade-off in rice. Nat Commun 14(1):4441. https://doi.org/10.1038/s41467-023-40176-2
Guihur A, Rebeaud ME, Goloubinoff P (2022) How do plants feel the heat and survive? Trends Biochem Sci 47(10):824–838. https://doi.org/10.1016/j.tibs.2022.05.004
Gurley WB (2000) HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell 12(4):457–460. https://doi.org/10.1105/tpc.12.4.457
Hahn A, Bublak D, Schleiff E, Scharf K-D (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23(2):741–755. https://doi.org/10.1105/tpc.110.076018
Haider S, Raza A, Iqbal J, Shaukat M, Mahmood T (2022) Analyzing the regulatory role of heat shock transcription factors in plant heat stress tolerance: a brief appraisal. Mol Biol Rep 49(6):5771–5785. https://doi.org/10.1007/s11033-022-07190-x
Hao L, Qiao X (2018) Genome-wide identification and analysis of the CNGC gene family in maize. PeerJ 6:e5816. https://doi.org/10.7717/peerj.5816
Hao D, Jin L, Wen X, Yu F, Xie Q, Guo H (2021) The RING E3 ligase SDIR1 destabilizes EBF1/EBF2 and modulates the ethylene response to ambient temperature fluctuations in Arabidopsis. Proc Natl Acad Sci 118(6):e2024592118. https://doi.org/10.1073/pnas.2024592118
He X, Guo S, Wang Y, Wang L, Shu S, Sun J (2020) Systematic identification and analysis of heat-stress-responsive lncRNAs, circRNAs and miRNAs with associated co-expression and ceRNA networks in cucumber (Cucumis sativus L.). Physiol Plant 168(3):736–754. https://doi.org/10.1111/ppl.12997
He K, Mei H, Zhu J, Qiu Q, Cao X, Deng X (2021) The histone H3K27 demethylase REF6/JMJ12 promotes thermomorphogenesis in Arabidopsis. Natl Sci Rev 9(5):nwab213. https://doi.org/10.1093/nsr/nwab213
He K, Cao X, Deng X (2021) Histone methylation in epigenetic regulation and temperature responses. Curr Opin Plant Biol 61:102001. https://doi.org/10.1016/j.pbi.2021.102001
Hu Z, Song N, Zheng M, Liu X, Liu Z, Xing J, Ma J, Guo W, Yao Y, Peng H, Xin M, Zhou DX, Ni Z, Sun Q et al (2015) Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J 84(6):1178–1191. https://doi.org/10.1111/tpj.13076
Hu X, Wei Q, Wu H, Huang Y, Peng X, Han G, Ma Q, Zhao Y (2022) Identification and characterization of heat-responsive lncRNAs in maize inbred line CM1. BMC Genomics 23(1):208. https://doi.org/10.1186/s12864-022-08448-1
Hu J, Zhao X, Gu L, Liu P, Zhao B, Zhang J, Ren B (2023) The effects of high temperature, drought, and their combined stresses on the photosynthesis and senescence of summer maize. Agric Water Manag 289:108525. https://doi.org/10.1016/j.agwat.2023.108525
Jagtap AB, Vikal Y, Johal GS (2020) Genome-wide development and validation of cost-effective KASP marker assays for genetic dissection of heat stress tolerance in maize. Int J Mol Sci 21(19):7386. https://doi.org/10.3390/ijms21197386
Jiang Y, Zheng Q, Chen L, Liang Y, Wu J (2017) Ectopic overexpression of maize heat shock transcription factor gene ZmHsf04 confers increased thermo and salt-stress tolerance in transgenic Arabidopsis. Acta Physiol Plant 40:9. https://doi.org/10.1007/s11738-017-2587-2
Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, Kumar M, Grant A, Locke JC, Schäfer E, Jaeger KE, Wigge PA (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354(6314):886–889. https://doi.org/10.1126/science.aaf6005
Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, Kim SB, Baek S, Zubieta C, Jaeger KE, Wigge PA (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585(7824):256–260. https://doi.org/10.1038/s41586-020-2644-7
Kan Y, Mu XR, Gao J, Lin HX, Lin Y (2023) The molecular basis of heat stress responses in plants. Mol Plant 16(10):1612–1634. https://doi.org/10.1016/j.molp.2023.09.013
Kimotho RN, Baillo EH, Zhang Z (2019) Transcription factors involved in abiotic stress responses in Maize (Zea mays L.) and their roles in enhanced productivity in the post genomics era. PeerJ 7:e7211. https://doi.org/10.7717/peerj.7211
Kohorn BD, Johansen S, Shishido A, Todorova T, Martinez R, Defeo E, Obregon P (2009) Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J 60:974–982. https://doi.org/10.1111/j.1365-313X.2009.04016.x
Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19(5):408–413. https://doi.org/10.1016/j.cub.2009.01.046
Kong W, Zhu Q, Zhang Q, Zhu Y, Yang J, Chai K, Lei W, Jiang M, Zhang S, Lin J, Zhang X (2023) 5mC DNA methylation modification-mediated regulation in tissue functional differentiation and important flavor substance synthesis of tea plant (Camellia sinensis L.). Hortic Res 10(8):uhad126. https://doi.org/10.1093/hr/uhad126
Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10(3):310–316. https://doi.org/10.1016/j.pbi.2007.04.011
Kraus K, Hnilickova H, Pecka J, Lhotska M, Bezdickova A, Martinek P, Kucirkova L, Hnilicka F (2022) The effect of the application of stimulants on the photosynthetic apparatus and the yield of winter wheat. Agronomy 12(1):78. https://doi.org/10.3390/agronomy12010078
Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484(7393):242–245. https://doi.org/10.1038/nature10928
Lämke J, Brzezinka K, Altmann S, Bäurle I (2016) A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 35(2):162-175. https://doi.org/10.15252/embj.201592593
Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11(3):204–220. https://doi.org/10.1038/nrg2719
Lee DG, Ahsan N, Lee SH, Kang KY, Bahk JD, Lee IJ, Lee BH (2007) A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 7(18):3369–3383. https://doi.org/10.1002/pmic.200700266
Li J, Liu J (2022a) TT3.1: a journey to protect chloroplasts upon heat stress. Stress Biol 2(1):27. https://doi.org/10.1007/s44154-022-00051-4
Li S, Liu J, Liu Z, Li X, Wu F, He Y (2014) Heat-induced TAS1 target1 mediates thermotolerance via heat stress transcription factor A1a–directed pathways in Arabidopsis. Plant Cell 26(4):1764–1780. https://doi.org/10.1105/tpc.114.124883
Li HC, Zhang HN, Li GL, Liu ZH, Zhang YM, Zhang HM, Guo XL (2015) Expression of maize heat shock transcription factor gene ZmHsf06 enhances the thermotolerance and drought-stress tolerance of transgenic Arabidopsis. Funct Plant Biol 42(11):1080–1091. https://doi.org/10.1071/FP15080
Li B, Gao K, Ren H, Tang W (2018) Molecular mechanisms governing plant responses to high temperatures. J Integr Plant Biol 60(9):757–779. https://doi.org/10.1111/jipb.12701
Li B, Gao Z, Liu X, Sun D, Tang W (2019a) Transcriptional profiling reveals a time-of-day-specific role of REVEILLE 4/8 in regulating the first wave of heat shock–induced gene expression in Arabidopsis. Plant Cell 31(10):2353–2369. https://doi.org/10.1105/tpc.19.00519
Li GL, Zhang HN, Shao H, Wang GY, Zhang YY, Zhang YJ, Zhao LN, Guo XL, Sheteiwy MS (2019b) ZmHsf05, a new heat shock transcription factor from Zea mays L. improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant. Plant Sci 283:375–384. https://doi.org/10.1016/j.plantsci.2019.03.002
Li Y, Li X, Yang J, He Y (2020a) Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat Commun 11(1):5351. https://doi.org/10.1038/s41467-020-19186-x
Li Z, Tang J, Srivastava R, Bassham DC, Howell SH (2020b) The transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. Plant Cell 32(11):3559–3575. https://doi.org/10.1105/tpc.20.00260
Li Y, Huang Y, Sun H, Wang T, Ru W, Pan L, Zhao X, Dong Z, Huang W, Jin W (2022c) Heat shock protein 101 contributes to the thermotolerance of male meiosis in maize. Plant Cell 34(10):3702–3717. https://doi.org/10.1093/plcell/koac184
Li J, Yang C, Xu J, Lu HP, Liu JX (2023a) The hot science in rice research: How rice plants cope with heat stress. Plant Cell Environ 46(4):1087–1103. https://doi.org/10.1111/pce.14509
Li J, Cao Y, Zhang J, Zhu C, Tang G, Yan J (2023b) The miR165/166-PHABULOSA module promotes thermotolerance by transcriptionally and posttranslationally regulating HSFA1. Plant Cell 35(8):2952–2971. https://doi.org/10.1093/plcell/koad121
Li Y, Ma H, Wu Y, Ma Y, Yang J, Li Y, Yue D, Zhang R, Kong J, Lindsey K, Zhang X, Min L (2023c) Single-cell transcriptome atlas and regulatory dynamics in developing cotton anthers. Adv Sci 17:e2304017. https://doi.org/10.1002/advs.202304017
Li B, Jiang S, Gao L, Wang W, Luo H, Dong Y, Gao Z, Zheng S, Liu X, Tang W (2024a) Heat shock factor A1s are required for phytochrome-interacting factor 4-mediated thermomorphogenesis in Arabidopsis. J Integr Plant Biol 66(1):20–35. https://doi.org/10.1111/jipb.13579
Li Z, Li Z, Ji Y, Wang C, Wang S, Shi Y, Le J, Zhang M (2024b) The ZmHSF20–ZmHSF4–ZmCesA2 module regulates heat stress tolerance in maize. bioRxiv. https://doi.org/10.1101/2024.02.21.581499
Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ (2011) Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12:76. https://doi.org/10.1186/1471-2164-12-76
Lin MY, Chai KH, Ko SS, Kuang LY, Lur HS, Charng YY (2014) A positive feedback loop between HEAT SHOCK PROTEIN101 and HEAT STRESS-ASSOCIATED 32-KD PROTEIN modulates long-term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol 164(4):2045–2053. https://doi.org/10.1104/pp.113.229609
Liu C, Lu F, Cui X, Cao X (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61:395–420. https://doi.org/10.1146/annurev.arplant.043008.091939
Liu Y, Zhang C, Chen J, Guo L, Li X, Li W, Yu Z, Deng J, Zhang P, Zhang K, Zhang L (2013) Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol Biochem 64:92–98. https://doi.org/10.1016/j.plaphy.2012.12.013
Liu J, Wang H, Chua N (2015) Long noncoding RNA transcriptome of plants. Plant Biotechnol J 13(3):319–328. https://doi.org/10.1111/pbi.12336
Liu HC, Lämke J, Lin SY, Hung MJ, Liu KM, Charng YY, Bäurle I (2018a) Distinct heat shock factors and chromatin modifications mediate the organ-autonomous transcriptional memory of heat stress. Plant J 95(3):401–413. https://doi.org/10.1111/tpj.13958
Liu J, Niu Y, Zhang J, Zhou Y, Ma Z, Huang X (2018b) Ca2+ channels and Ca2+ signals involved in abiotic stress responses in plant cells: recent advances. Plant Cell Tissue Organ Cult 132:413–424. https://doi.org/10.1007/s11240-017-1350-0
Liu Q, Galli M, Liu X, Federici S, Buck A, Cody J, Labra M, Gallavotti A (2019a) NEEDLE1 encodes a mitochondria localized ATP-dependent metalloprotease required for thermotolerant maize growth. Proc Natl Acad Sci 116(39):19736–19742. https://doi.org/10.1073/pnas.1907071116
Liu J, Feng L, Gu X, Deng X, Qiu Q, Li Q, Zhang Y, Wang M, Deng Y, Wang E, He Y, Bäurle I, Li J, Cao X, He Z (2019b) An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res 29(5):379–390. https://doi.org/10.1038/s41422-019-0145-8
Liu P, Liu R, Xu Y, Zhang C, Niu Q, Lang Z (2023a) DNA cytosine methylation dynamics and functional roles in horticultural crops. Hortic Res 10(10):uhad170. https://doi.org/10.1093/hr/uhad170
Liu L, Zhang Y, Tang C, Shen Q, Fu J, Wang Q (2023b) Maize transcription factor ZmHsf28 positively regulates plant drought tolerance. Int J Mol Sci. 24(9):8079. https://doi.org/10.3390/ijms24098079
Lobell DB, Bänziger M, Magorokosho C, Vivek B (2011) Nonlinear heat effects on African maize as evidenced by historical yield trials. Nat Clim Change 1:42–45. https://doi.org/10.1038/nclimate1043
Matthews C, Arshad M, Hannoufa A (2019) Alfalfa response to heat stress is modulated by microRNA156. Physiol Plant 165:830–842. https://doi.org/10.1111/ppl.12787
Mittler R, Finka A, Goloubinoff P (2011) How do plants feel the heat? Trends Biochem Sci 37(3):118–25. https://doi.org/10.1016/j.tibs.2011.11.007
Navarro JA, Saiz-Bonilla M, Sanchez-Navarro JA, Pallas V (2021) The mitochondrial and chloroplast dual targeting of a multifunctional plant viral protein modulates chloroplast-to-nucleus communication, RNA silencing suppressor activity, encapsidation, pathogenesis and tissue tropism. Plant J 108(1):197–218. https://doi.org/10.1111/tpj.15435
Niu Y, Bai J, Liu X, Zhang H, Bao J, Zhao W, Hou Y, Deng X, Yang C, Guo L et al (2022) HISTONE DEACETYLASE 9 transduces heat signal in plant cells. Proc Natl Acad Sci U S A 119(45):e2206846119. https://doi.org/10.1073/pnas.2206846119
Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22(1):53–65. https://doi.org/10.1016/j.tplants.2016.08.015
Perrella G, Bäurle I, van Zanten M (2022) Epigenetic regulation of thermomorphogenesis and heat stress tolerance. New Phytol 234(4):1144–1160. https://doi.org/10.1111/nph.17970
Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LS, Shinozaki K, Yamaguchi-Shinozaki K (2007) Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J 50(1):54–69. https://doi.org/10.1111/j.1365-313X.2007.03034.x
Qin Q, Zhao Y, Zhang J, Chen L, Si W, Jiang H (2022) A maize heat shock factor ZmHsf11 negatively regulates heat stress tolerance in transgenic plants. BMC Plant Biol 22(1):406. https://doi.org/10.1186/s12870-022-03789-1
Rytz TC, Miller MJ, McLoughlin F, Augustine RC, Marshall RS, Juan YT, Charng YY, Scalf M, Smith LM, Vierstra RD (2018) SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell 30(5):1077–1099. https://doi.org/10.1105/tpc.17.00993
Sánchez B, Rasmussen A, Porter JR (2014) Temperatures and the growth and development of maize and rice: a review. Glob Change Biol 20(2):408–417. https://doi.org/10.1111/gcb.12389
Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P (2008) A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J Cell Mol Biol 53(2):264–274. https://doi.org/10.1111/j.1365-313X.2007.03334.x
Sharma M, Kumar P, Verma V, Sharma R, Bhargava B, Irfan M (2022) Understanding plant stress memory response for abiotic stress resilience: Molecular insights and prospects. Plant Physiol Biochem 179:10–24. https://doi.org/10.1016/j.plaphy.2022.03.004
Shekhawat K, Saad MM, Sheikh A, Mariappan K, Al-Mahmoudi H, Abdulhakim F, Eida AA, Jalal R, Masmoudi K, Hirt H (2021) Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep 22(3):e51049. https://doi.org/10.15252/embr.202051049
Shen X, De Jonge J, Forsberg SK, Pettersson ME, Sheng Z, Hennig L, Carlborg Ö (2014) Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PloS Genet 10(12):e1004842. https://doi.org/10.1371/journal.pgen.1004842
Shen Y, Lei T, Cui X, Liu X, Zhou S, Zheng Y, Guérard F, Issakidis-Bourguet E, Zhou DX (2019) Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J 100(5):991-1006. https://doi.org/10.1111/tpj.14492.
Singh A, Pandey H, Pandey S, Lal D, Chauhan D, Aparna Antre SHBS, Kumar A (2023) Drought stress in maize: stress perception to molecular response and strategies for its improvement. Funct Integr Genomics 23(4):296. https://doi.org/10.1007/s10142-023-01226-6
Song Y, Chen P, Liu P, Bu C, Zhang D (2020) High-Temperature-Responsive poplar lncRNAs modulate target gene expression via RNA interference and act as RNA scaffolds to enhance heat tolerance. Int J Mol Sci 21(18):6808. https://doi.org/10.3390/ijms21186808
Song ZT, Zhang LL, Han JJ, Zhou M, Liu JX (2021) Histone H3K4 methyltransferases SDG25 and ATX1 maintain heat-stress gene expression during recovery in Arabidopsis. Plant J 105(5):1326–1338. https://doi.org/10.1111/tpj.15114
Sun L, Wen J, Peng H, Yao Y, Hu Z, Ni Z, Sun Q, Xin M (2021) The genetic and molecular basis for improving heat stress tolerance in wheat. aBIOTECH 3(1):25–39. https://doi.org/10.1007/s42994-021-00064-z
Sunkar R, Li Y-F, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17(4):196–203. https://doi.org/10.1016/j.tplants.2012.01.010
Tan W, Chen J, Yue X, Chai S, Liu W, Li C, Yang F, Gao Y, Gutiérrez Rodríguez L, Resco de Dios V, Zhang D, Yao Y (2023) The heat response regulators HSFA1s promote Arabidopsis thermomorphogenesis via stabilizing PIF4 during the day. Sci Adv 9(44):eadh1738. https://doi.org/10.1126/sciadv.adh1738
Ueda M, Seki M (2020) Histone modifications form epigenetic regulatory networks to regulate abiotic stress response1. Plant Physiol 182(1):15–26. https://doi.org/10.1104/pp.19.00988
van der Woude LC, Perrella G, Snoek BL, van Hoogdalem M, Novák O, van Verk MC, van Kooten HN, Zorn LE, Tonckens R, Dongus JA et al (2019) HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc Natl Acad Sci 116(50):25343–25354. https://doi.org/10.1073/pnas.1911694116
Vu LD, Gevaert K, De Smet I (2019) Feeling the heat: searching for plant thermosensors. Trends Plant Sci 24(3):210–219. https://doi.org/10.1016/j.tplants.2018.11.004
Wan J, He M, Hou Q, Zou L, Yang Y, Wei Y, Chen X (2021) Cell wall associated immunity in plants. Stress Biol 1(1):3. https://doi.org/10.1007/s44154-021-00003-4
Wang H, Niu H, Liang M, Zhai Y, Huang W, Ding Q, Du Y, Lu M (2019) A wall-associated kinase gene CaWAKL20 from pepper negatively modulates plant thermotolerance by reducing the expression of ABA-responsive genes. Front Plant Sci 10:591. https://doi.org/10.3389/fpls.2019.00591
Wang J, Chen L, Long Y, Si W, Cheng B, Jiang H (2021) A novel heat shock transcription factor (ZmHsf08) negatively regulates salt and drought stress responses in maize. Int J Mol Sci 22(21):11922. https://doi.org/10.3390/ijms222111922
Wang X, Han L, Li J, Shang X, Liu Q, Li L, Zhang H (2023) Next-generation bulked segregant analysis for Breeding 4.0. Cell Rep 42:113039. https://doi.org/10.1016/j.celrep.2023.113039
Wang H, Feng M, Jiang Y, Du D, Dong C, Zhang Z, Wang W, Liu J, Liu X, Li S, Chen Y, Guo W, Xin M, Yao Y, Ni Z, Sun Q, Peng H, Liu J (2023) Thermosensitive SUMOylation of TaHsfA1 defines a dynamic ON/OFF molecular switch for the heat stress response in wheat. Plant Cell 35(10):3889–3910. https://doi.org/10.1093/plcell/koad192
Wolf S (2022) Cell wall signaling in plant development and defense. Annu Rev Plant Biol 73:323–353. https://doi.org/10.1146/annurev-arplant-102820-095312
Wu HC, Bulgakov VP, Jinn TL (2018) Pectin methylesterases: cell wall remodeling proteins are required for plant response to heat stress. Front Plant Sci 9:1612. https://doi.org/10.3389/fpls.2018.01612
Wu L, Han L, Li Q, Wang G, Zhang H, Li L (2021a) Using interactome big data to crack genetic mysteries and enhance future crop breeding. Mol Plant 14(4):77–94. https://doi.org/10.1016/j.molp.2020.12.012
Wu L, Luo Z, Shi Y, Jiang Y, Li R, Miao X, Yang F, Li Q, Zhao H, Xue J, Xu S, Zhang T, Li L (2021b) A cost-effective tsCUT&Tag method for profiling transcription factor binding landscape. J Integr Plant Biol 64(11):2033–2038. https://doi.org/10.1111/jipb.13354
Wu Z, Li T, Xiang J, Teng R, Zhang D, Teng N (2023) A lily membrane-associated NAC transcription factor, LlNAC014, is involved in thermotolerance via activation of the DREB2-HSFA3 module. J Exp Bot 74(3):945–963. https://doi.org/10.1093/jxb/erac436
Wu Z, Li T, Ding L, Wang C, Teng R, Xu S, Cao X, Teng N (2024) Lily LlHSFC2 coordinates with HSFAs to balance heat stress response and improve thermotolerance. New Phytol 241(5):2124–2142. https://doi.org/10.1111/nph.19507
Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M (2007) Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem 282(52):37794–37804. https://doi.org/10.1074/jbc.M707168200
Yamaguchi N, Matsubara S, Yoshimizu K, Seki M, Hamada K, Kamitani M, Kurita Y, Nomura Y, Nagashima K, Inagaki S, Suzuki T, Gan ES, To T, Kakutani T, Nagano AJ, Satake A, Ito T (2021) H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat Commun 12(1):3480. https://doi.org/10.1038/s41467-021-23766-w
Yang Y, Saand MA, Huang L, Abdelaal WB, Zhang J, Wu Y, Li J, Sirohi MH, Wang F (2021) Applications of multi-omics technologies for crop improvement. Front Plant Sci 12:563953. https://doi.org/10.3389/fpls.2021.563953
Yang Z, Cao Y, Shi Y, Qin F, Jiang C, Yang S (2023) Genetic and molecular exploration of maize environmental stress resilience: toward sustainable agriculture. Mol Plant 16(10):1496–1517. https://doi.org/10.1016/j.molp.2023.07.005
Yao X, Li Y, Chen J, Zhou Z, Wen Y, Fang K, Yang F, Li T, Zhang D, Lin H (2022) Brassinosteroids enhance BES1-required thermomemory in Arabidopsis thaliana. Plant Cell Environ 45(12):3492–3504. https://doi.org/10.1111/pce.14444
Yin Y, Qin K, Song X, Zhang Q, Zhou Y, Xia X, Yu J (2018) BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol 59(11):2239–2254. https://doi.org/10.1093/pcp/pcy146
Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim JM, Seki M, Todaka D, Osakabe Y, Sakuma Y, Schöffl F, Shinozaki K, Yamaguchi-Shinozaki K (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286(5–6):321–332. https://doi.org/10.1007/s00438-011-0647-7
Yu X, Liu Z, Sun X (2023) Single-cell and spatial multi-omics in the plant sciences: Technical advances, applications, and perspectives. Plant Commun 4(3):100508. https://doi.org/10.1016/j.xplc.2022.100508
Zhang H, Li G, Fu C, Duan S, Hu D, Guo X (2020a) Genome-wide identification, transcriptome analysis and alternative splicing events of Hsf family genes in maize. Sci Rep 10(1):8073. https://doi.org/10.1038/s41598-020-65068-z
Zhang H, Li G, Hu D, Zhang Y, Zhang Y, Shao H, Zhao L, Yang R, Guo X (2020b) Functional characterization of maize heat shock transcription factor gene ZmHsf01 in thermotolerance. PeerJ 8:e8926. https://doi.org/10.7717/peerj.8926
Zhang H, Zhou JF, Kan Y, Shan JX, Ye WW, Dong NQ, Guo T, Xiang YH, Yang YB, Li YC et al (2022) A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 376(6599):1293–1300. https://doi.org/10.1126/science.abo5721
Zhang HN, Meng XZ, Li R, Ma ZY, Liu R, Liu ZH, Duan SN, Zhang WT, Li GL, Guo X (2024) Intron retention via alternative splicing affects the thermotolerance regulation of ZmHsf17. Physiol Plant 176:e14138. https://doi.org/10.1111/ppl.14138
Zhao H, Bao Y (2021) PIF4: Integrator of light and temperature cues in plant growth. Plant Sci 313:111086. https://doi.org/10.1016/j.plantsci.2021.111086
Zhao C, Liu B, Piao S, Wang X, Lobell DB, Huang Y, Huang M, Yao Y, Bassu S, Ciais P et al (2017) Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci 114(35):9326–9331. https://doi.org/10.1073/pnas.1701762114
Zhao Y, Du H, Wang Y, Wang H, Yang S, Li C, Chen N, Yang H, Zhang Y, Zhu Y, Yang L, Hu X (2021) The calcium-dependent protein kinase ZmCDPK7 functions in heat-stress tolerance in maize. J Integr Plant Biol 63(3):510–527. https://doi.org/10.1111/jipb.13056
Zhao Y, Wei Q, Chen T, Xu L, Liu J, Zhang X, Han G, Ma Q (2022) Identification and characterization of heat-responsive miRNAs and their regulatory network in maize. Plant Growth Regul 96:195–208. https://doi.org/10.1007/s10725-021-00769-7
Zhu J, Cao X, Deng X (2023) Epigenetic and transcription factors synergistically promote the high temperature response in plants. Trends Biochem Sci 48(9):788–800. https://doi.org/10.1016/j.tibs.2023.06.001
Acknowledgements
Not applicable.
Funding
This work was supported by grants from Program for High-level Talents Recruitment of Anhui Agricultural University (rc422208) and Anhui Agricultural University fund (rc312212).
Author information
Authors and Affiliations
Contributions
This work was supervised by Leiming Wu. Yong-Ling Ruan made intellectual contributions to the writing and revision of the manuscript. The original draft was prepared by Mingxiu Ruan and Heng Zhao. Yujing Wen, Hao Chen, Feng He and Xiaoqin Song collected related publications and revised the manuscript. Xinbo Hou helped to draw and revise the figures. Haiyang Jiang improved the manuscript. All author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agree to publish.
Competing interests
All authors declare no competing interests.
Additional information
Handling Editor: Dr. Huazhong Shi.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ruan, M., Zhao, H., Wen, Y. et al. The complex transcriptional regulation of heat stress response in maize. Stress Biology 4, 24 (2024). https://doi.org/10.1007/s44154-024-00165-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44154-024-00165-x