Abstract
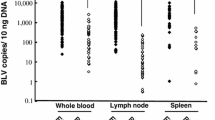
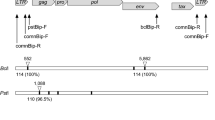
Bovine leukemia virus (BLV) is an oncogenic member of the genus Deltaretrovirus. BLV infects cattle worldwide and is responsible for significant economic losses. The objective of this study was to validate real-time quantitative PCR (qPCR) for the detection of BLV. After identification of the most efficient qPCR, the limits of detection, repeatability, and reproducibility were determined. The results indicate that qPCR can be easily reproduced between laboratories with high sensitivity. The test variation was low in samples from lesions suggestive of bovine leukosis or whole blood.
Similar content being viewed by others
Data availability
Data are available upon request.
References
Kettmann R, Portetelle D, Mammerickx M, Cleuter Y, Dekegel D, Galoux M, Ghysdael J, Burny A, Chantrenne H (1976) Bovine leukemia virus: an exogenous RNA oncogenic virus. Proc Natl Acad Sci USA 73(4):1014–1018
Nekouei O, VanLeeuwen J, Stryhn H, Kelton D, Keefe G (2016) Lifetime effects of infection with bovine leukemia virus on longevity and milk production of dairy cows. Prev Vet Med 133:1–9
Delarmelina E, Buzelin MA, Souza BS, Souto FM, Bicalho JM, Câmara RJF, Resende CF, Bueno BL, Victor RM, Galinari GCF, Nunes CB, Leite RC, Costa ÉA, Reis JKPD (2020) High positivity values for bovine leukemia virus in human breast cancer cases from Minas Gerais, Brazil. PLOS One 15(10):e0239745
Murakami H, Uchiyama J, Suzuki C, Nikaido S, Shibuya K, Sato R, Maeda Y, Tomioka M, Takeshima SN, Kato H, Sakaguchi M, Sentsui H, Aida Y, Tsukamoto K (2018) Variations in the viral genome and biological properties of bovine leukemia virus wild-type strains. Virus Res 253:103–111. https://doi.org/10.1016/j.virusres.2018.06.005
Camargos MF, Stancek D, Rocha MA, Lessa LM, Reis JK, Leite RC (2002) Partial sequencing of env gene of bovine leukaemia virus from Brazilian samples and phylogenetic analysis. J Vet Med B Infect Dis Vet Public Health 49(7):325–331
Yu C, Wang X, Zhou Y, Wang Y, Zhang X, Zheng Y (2019) Genotyping bovine leukemia virus in dairy cattle of Heilongjiang, northeastern China. BMC Vet Res 15(1):179
Hirsch C, Camargos MF, Barbosa-Stancioli EF et al (2015) Genetic variability and phylogeny of the 5′ long terminal repeat from Brazilian bovine leukemia virus. Genet Mol Res 14(4):14530–14538
Jimba M, Takeshima SN, Murakami H et al (2012) BLV-CoCoMo-qPCR: a useful tool for evaluating bovine leukemia virus infection status. BMC Vet Res 8:167
Naif HM, Brandon RB, Daniel RCW, Lavin MF (1990) Bovine leukemia proviral DNA detection in cattle using the polymerase chain-reaction. Vet Microbiol 25(2–3):117–129
Burridge MJ, Thurmond MC, Miller JM, Schmerr MJ, Van Der Maaten MJ (1982) Fall in antibody titer to bovine leukemia virus in the periparturient period. Can J Comp Med 46(3):270–271
Dias NL, Fonseca Júnior AA, Rodrigues DS, Camargos MF (2012) PCR em tempo real para diagnóstico da leucose enzoótica bovina. Cien Rural 42(8):1434–1439
Miyasaka T, Takeshima SN, Jimba M, Matsumoto Y, Kobayashi N, Matsuhashi T, Sentsui H, Aida Y (2013) Identification of bovine leukocyte antigen class II haplotypes associated with variations in bovine leukemia virus proviral load in Japanese Black cattle. Tissue Antigens 81(2):72–82
OIE (2019) Manual of diagnostic tests and vaccines for terrestrial animals: Chapter 2.4.11. Enzootic Bovine Leukosis, 7th edn. World organization for animal health, France
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35(Web Server issue):W71–W74
Pinheiro de Oliveira TF, Fonseca AA, Camargos MF et al (2013) Detection of contaminants in cell cultures, sera and trypsin. Biologicals 41(6):407–414
Klintevall K, Ballagi-Pordány A, Näslund K, Belák S (1994) Bovine leukaemia virus: rapid detection of proviral DNA by nested PCR in blood and organs of experimentally infected calves. Vet Microbiol 42(2–3):191–204
de Oliveira AM, Fonseca AA, Camargos MF et al (2018) Development and validation of rt-qpcr for vesicular stomatitis virus detection (Alagoas Vesiculovirus). J Virol Methods 257:7–11
Barez PY, de Brogniez A, Carpentier A et al (2015) Recent advances in BLV research. Viruses 7(11):6080–6088
Bustin S, Huggett J (2017) qPCR primer design revisited. Biomol Detect Quantif 14:19–28
Jimba M, Takeshima SN, Matoba K et al (2010) BLV-CoCoMo-qPCR: quantitation of bovine leukemia virus proviral load using the CoCoMo algorithm. Retrovirology 7:91
Taylor GM, Worth DR, Palmer S, Jahans K, Hewinson RG (2007) Rapid detection of Mycobacterium bovis DNA in cattle lymph nodes with visible lesions using PCR. BMC Vet Res 3:12
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622
Stenman J, Orpana A (2001) Accuracy in amplification. Nat Biotechnol 19(11):1011–1012
Funding
This work was supported by the Ministério da Agricultura, Pecuária e Abastecimento, and Universidade Federal de Minas Gerais.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Jorge Luiz Mello Sampaio
Rights and permissions
About this article
Cite this article
Fonseca Júnior, A.A., Ferreira, L.R., Laguardia- Nascimento, M. et al. Evaluation of three different genomic regions for detection of bovine leukemia virus by real-time PCR. Braz J Microbiol 52, 2483–2488 (2021). https://doi.org/10.1007/s42770-021-00613-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00613-0