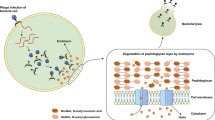
Abstract
Staphylococcus aureus is the leading cause of secondary infections in hospitals and a challenging pathogen in food industries. Decades after it was first reported, β-lactam-resistant S. aureus remains a subject of intense research owing to the ever-increasing issue of drug resistance. S. aureus bacteriophages (phages) or their encoded products are considered an alternative to antibiotics as they have been shown to be effective in treating some S. aureus-associated infections. In this review, we present a concise collection of the literature on the pathogenic potential of S. aureus and examine the prospects of using S. aureus phages and their encoded products as antimicrobials.

Similar content being viewed by others
References
Lowy FD (1998) Staphylococcus aureus infections. N Engl J Med 339(8):520–532. https://doi.org/10.1056/NEJM199808203390806
Williams REO (1963) Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev 27(1):56
Wertheim HF, Melles DC, Vos MC, Van Leeuwen W, Van Belkum A, Verbrugh HA, Nouwen JL (2005) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5(12):751–762. https://doi.org/10.1016/S1473-3099(05)70295-4
Taylor TA, Unakal CG (2020) Staphylococcus aureus. [Updated 2020 Aug 23]. In: StatPearls. StatPearls Publishing, Treasure Island (FL). Available from: https://www.ncbi.nlm.nih.gov/books/NBK441868/
Foster T (1996) Staphylococcus. In: Baron S (ed) Medical Microbiology, 4th edn. The University of Texas Medical Branch, Galveston, pp 187–197
Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR et al (2015) Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog 11(12):e1005341. https://doi.org/10.1371/journal.ppat.1005341
McGuinness WA, Malachowa N, DeLeo FR (2017) Focus: infectious diseases: vancomycin-resistance in Staphylococcus aureus. Yale J Biol Med 90(2):269–281
WHO (2020) Global action plan on AMR https://www.who.int/antimicrobial-resistance/global-action-plan/en/ accessed 29 January 2020
Úbeda C, Maiques E, Knecht E, Lasa Í, Novick RP, Penadés JR (2005) Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol 56(3):836–844. https://doi.org/10.1111/j.1365-2958.2005.04584.x
Singh R, Sripada L, Singh R (2014) Side effects of antibiotics during bacterial infection: mitochondria, the main target in host cell. Mitochondrion 16:50–54. https://doi.org/10.1016/j.mito.2013.10.005
Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB (2014) Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 68:217–235. https://doi.org/10.1146/annurev-micro-091313-103456
Pantosti A, Sanchini A, Monaco M (2007) Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol 2:323–334. https://doi.org/10.2217/17460913.2.3.323
CDC (2019) Vital signs www.cdc.gov/vitalsigns/staph/index.html accessed on 05 April 2021
Diekema DJ, Pfaller MA, Shortridge D, Zervos M, Jones RN (2019) Twenty-year trends in antimicrobial susceptibilities among Staphylococcus aureus from the SENTRY Antimicrobial Surveillance Program. Open Forum Infect Dis 6(Suppl 1):S47–S53. https://doi.org/10.1093/ofid/ofy270
www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance accessed on 05 April 2021
Furfaro LL, Payne MS, Chang BJ (2018) Bacteriophage therapy: clinical trials and regulatory hurdles. Front Cell Infect Microbiol 8:376. https://doi.org/10.3389/fcimb.2018.00376
Merril CR, Scholl D, Adhya SL (2003) The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2(6):489–497. https://doi.org/10.1038/nrd1111
Deghorain M, Van Melderen L (2012) The Staphylococci phages family: an overview. Viruses 4(12):3316–3335. https://doi.org/10.3390/v4123316
Lyon BR, Skurray RON (1987) Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev 51(1):88–134
Morikawa K, Takemura AJ, Inose Y, Tsai M, Ohta T, Msadek T (2012) Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog 8(11):e1003003. https://doi.org/10.1371/journal.ppat.1003003
Everitt RG, Didelot X, Batty EM, Miller RR, Knox K, Young BC, Bowden R, Auton A, Votintseva A, Larner-Svensson H, Charlesworth J (2014) Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nat Commun 5(1):1–9. https://doi.org/10.1038/ncomms4956
Alibayov B, Zdenkova K, Sykorova H, Demnerova K (2014) Molecular analysis of Staphylococcus aureus pathogenicity islands (SaPI) and their superantigens combination of food samples. J Microbiol Methods 107:197–204. https://doi.org/10.1016/j.mimet.2014.10.014
Grundstad ML, Parlet CP, Kwiecinski JM, Kavanaugh JS, Crosby HA, Cho YS, Heilmann K, Diekema DJ, Horswill AR (2019) Quorum sensing, virulence, and antibiotic resistance of USA100 methicillin-resistant Staphylococcus aureus isolates. mSphere 4(4):e00553-19. https://doi.org/10.1128/mSphere.00553-19
Jarraud S, Lyon GJ, Figueiredo AMS, Lina G, Vandenesch F, Etienne J, Muir TW, Novick RP (2011) Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol 193(24):7027. https://doi.org/10.1128/JB.06355-11
Cheung AL, Nishina KA, Trotonda MP, Tamber S (2008) The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol 40(3):355–361. https://doi.org/10.1016/j.biocel.2007.10.032
Pichon C, Felden B (2005) Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. PNAS 102(40):14249–14254. https://doi.org/10.1073/pnas.0503838102
Desgranges E, Marzi K, Moreau K, Romby P, Caldelari I (2019) Chapter 35 Noncoding RNA. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Braunstein M, Rood JI (eds) Gram-positive Pathogens, 3rd edn. Wiley, New York
Haaber J, Penadés JR, Ingmer H (2017) Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol 25(11):893–905. https://doi.org/10.1016/j.tim.2017.05.011
Deurenberg RH, Stobberingh EE (2008) The evolution of Staphylococcus aureus. Infect Genet Evol 8(6):747–763. https://doi.org/10.1016/j.meegid.2008.07.007
Baig S, Johannesen TB, Overballe-Petersen S, Larsen J, Larsen AR, Stegger M (2018) Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect Genet Evol 61:74–76. https://doi.org/10.1016/j.meegid.2018.03.013
David MZ, Daum RS (2010) Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23(3):616–687. https://doi.org/10.1128/cmr.00081-09
Boucher HW, Corey GR (2008) Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis 46(Supplement 5):S344–S349. https://doi.org/10.1086/533590
Pantosti A (2012) Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front Microbiol 3:127. https://doi.org/10.3389/fmicb.2012.00127
Verstappen KM, Tulinski P, Duim B, Fluit AC, Carney J, Van Nes A, Wagenaar JA (2016) The effectiveness of bacteriophages against methicillin-resistant Staphylococcus aureus ST398 nasal colonization in pigs. PLoS ONE 11(8):e0160242. https://doi.org/10.1371/journal.pone.0160242
Figueiredo AMS, Ferreira FA (2014) The multifaceted resources and microevolution of the successful human and animal pathogen methicillin-resistant Staphylococcus aureus. Mem Inst Oswaldo Cruz 109(3):265–278. https://doi.org/10.1590/0074-0276140016
Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS (2008) Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis 198(4):561–570. https://doi.org/10.1086/590157
Xia G, Wolz C (2014) Phages of Staphylococcus aureus and their impact on host evolution. Infect Genet Evol 21:593–601. https://doi.org/10.1016/j.meegid.2013.04.022
Kwan T, Liu J, DuBow M, Gros P, Pelletier J (2005) The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci USA 102(14):5174–5179. https://doi.org/10.1073/pnas.0501140102
Oliveira H, Sampaio M, Melo LD, Dias O, Pope WH, Hatfull GF, Azeredo J (2019) Staphylococci phages display vast genomic diversity and evolutionary relationships. BMC Genomics 20(1):357. https://doi.org/10.1186/s12864-019-5647-8
O’Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP (2004) Genome of staphylococcal phage K: a new lineage of Myoviridae infecting Gram-positive bacteria with a low G+ C content. J Bacteriol 186(9):2862–2871. https://doi.org/10.1128/JB.186.9.2862-2871.2004
Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Bröker BM, Doskar J, Wolz C (2009) Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol 191(11):3462–3468. https://doi.org/10.1128/jb.01804-08
Chang Y, Shin H, Lee JH, Park CJ, Paik SY, Ryu S (2015) Isolation and genome characterization of the virulent Staphylococcus aureus bacteriophage SA97. Viruses 7(10):5225–5242. https://doi.org/10.3390/v7102870
Głowacka-Rutkowska A, Gozdek A, Empel J, Gawor J, Żuchniewicz K, Kozińska A, Dębski J, Gromadka R, Łobocka M (2019) The ability of lytic staphylococcal podovirus vB_SauP_phiAGO1. 3 to coexist in equilibrium with its host facilitates the selection of host mutants of attenuated virulence but does not preclude the phage antistaphylococcal activity in a nematode infection model. Front Microbiol 9:3227. https://doi.org/10.3389/fmicb.2018.03227
Rakhuba DV, Kolomiets EI, Dey ES, Novik GI (2010) Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol 59(3):145–155
Winstel V, Xia G, Peschel A (2014) Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int J Med Microbiol 304(3–4):215–221. https://doi.org/10.1016/j.ijmm.2013.10.009
Azam AH, Tanji Y (2019) Peculiarities of Staphylococcus aureus phages and their possible application in phage therapy. Appl Microbiol Biotechnol 103:4279–4289. https://doi.org/10.1007/s00253-019-09810-2
Koç C, Xia G, Kühner P, Spinelli S, Roussel A, Cambillau C, Stehle T (2016) Structure of the host-recognition device of Staphylococcus aureus phage ϕ11. Sci Rep 6(1):1–11. https://doi.org/10.1038/srep27581
Takeuchi I, Osada K, Azam AH, Asakawa H, Miyanaga K, Tanji Y (2016) The presence of two receptor-binding proteins contributes to the wide host range of staphylococcal Twort-like phages. Appl Environ Microbiol 82(19):5763–5774. https://doi.org/10.1128/AEM.01385-16
Cui Z, Guo X, Dong K, Zhang Y, Li Q, Zhu Y, Zeng L, Tang R, Li L (2017) Safety assessment of Staphylococcus phages of the family Myoviridae based on complete genome sequences. Sci Rep 7(1):1–8. https://doi.org/10.1038/srep41259
Ajuebor J, Buttimer C, Arroyo-Moreno S, Chanishvili N, Gabriel EM, O’Mahony J, McAuliffe O, Neve H, Franz C, Coffey A (2018) Comparison of Staphylococcus phage K with close phage relatives commonly employed in phage therapeutics. Antibiotics 7(2):37. https://doi.org/10.3390/antibiotics7020037
Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, Glonti T, Krylov V, Mast J, Van Parys L, Lavigne R (2009) Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE 4(3):e4944. https://doi.org/10.1371/journal.pone.0004944
Sulakvelidze A, Kutter E (2005) Bacteriophage therapy. In: Kutter E, Sulakvelidze A (eds) Bacteriophage biology and applications, 1st edn. CRC Press, Boca Raton, p 381–436
Van Belleghem JD, Clement F, Merabishvili M, Lavigne R, Vaneechoutte M (2017) Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci Rep 7(1):8004. https://doi.org/10.1038/s41598-017-08336-9
Gutiérrez D, Fernández L, Rodríguez A, García P (2018) Are phage lytic proteins the secret weapon to kill Staphylococcus aureus? MBio 9(1):e01923-e2017. https://doi.org/10.1128/mBio.01923-17
Adhya S, Merril CR, Biswas B (2014) Therapeutic and prophylactic applications of bacteriophage components in modern medicine. Cold Spring Harb Perspect Biol 4(1):a012518. https://doi.org/10.1101/cshperspect.a012518
Schmelcher M, Shen Y, Nelson DC, Eugster MR, Eichenseher F, Hanke DC, Loessner MJ, Dong S, Pritchard DG, Lee JC, Becker SC (2015) Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. J Antimicrob Chemother 70(5):1453–1465. https://doi.org/10.1093/jac/dku552
Love MJ, Bhandari D, Dobson RC, Billington C (2018) Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics 7(1):17. https://doi.org/10.3390/antibiotics7010017
Becker SC, Roach DR, Chauhan VS, Shen Y, Foster-Frey J, Powell AM, Bauchan G, Lease RA, Mohammadi H, Harty WJ, Simmons C (2016) Triple-acting lytic enzyme treatment of drug-resistant and intracellular Staphylococcus aureus. Sci Rep 6(1):1–10. https://doi.org/10.1038/srep25063
Becker SC, Swift S, Korobova O, Schischkova N, Kopylov P, Donovan DM, Abaev I (2015) Lytic activity of the staphylolytic Twort phage endolysin CHAP domain is enhanced by the SH3b cell wall binding domain. FEMS Microbiol Lett 362(1):1–8. https://doi.org/10.1093/femsle/fnu019
Sass P, Bierbaum G (2007) Lytic activity of recombinant bacteriophage φ11 and φ12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl Environ Microbiol 73(1):347–352. https://doi.org/10.1128/AEM.01616-06
Chang Y, Yoon H, Kang DH, Chang PS, Ryu S (2017) Endolysin LysSA97 is synergistic with carvacrol in controlling Staphylococcus aureus in foods. Int J Food Microbiol 244:19–26. https://doi.org/10.1016/j.ijfoodmicro.2016.12.007
Zhou Y, Zhang H, Bao H, Wang X, Wang R (2017) The lytic activity of recombinant phage lysin LysKΔamidase against staphylococcal strains associated with bovine and human infections in the Jiangsu province of China. Res Vet Sci 111:113–119. https://doi.org/10.1016/j.rvsc.2017.02.011
Obeso JM, Martínez B, Rodríguez A, García P (2008) Lytic activity of the recombinant staphylococcal bacteriophage ΦH5 endolysin active against Staphylococcus aureus in milk. Int J Food Microbiol 128(2):212–218. https://doi.org/10.1016/j.ijfoodmicro.2008.08.010
Fenton M, Keary R, McAuliffe O, Ross RP, O’Mahony J, Coffey A (2013) Bacteriophage-derived peptidase CHAP (K) eliminates and prevents Staphylococcal biofilms. Int J Microbiol 2013:625341. https://doi.org/10.1155/2013/625341
Schmelcher M, Powell AM, Becker SC, Camp MJ, Donovan DM (2012) Chimeric phage lysins act synergistically with lysostaphin to kill mastitis-causing Staphylococcus aureus in murine mammary glands. Appl Environ Microbiol 78(7):2297–2305. https://doi.org/10.1128/aem.07050-11
Takáč M, Bläsi U (2005) Phage P68 virion-associated protein 17 displays activity against clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother 49(7):2934–2940. https://doi.org/10.1128/AAC.49.7.2934-2940.2005
Paul VD, Rajagopalan SS, Sundarrajan S, George SE, Asrani JY, Pillai R, Chikkamadaiah R, Durgaiah M, Sriram B, Padmanabhan S (2011) A novel bacteriophage Tail-Associated Muralytic Enzyme (TAME) from Phage K and its development into a potent antistaphylococcal protein. BMC Microbiol 11(1):1–11. https://doi.org/10.1186/1471-2180-11-226
Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T et al (2004) Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol 22:185–191. https://doi.org/10.1038/nbt932
Kashani HH, Schmelcher M, Sabzalipoor H, Hosseini ES, Moniri R (2018) Recombinant endolysins as potential therapeutics against antibiotic-resistant Staphylococcus aureus: current status of research and novel delivery strategies. Clin Microbiol Rev 31(1):e00071-17. https://doi.org/10.1128/CMR.00071-17
Dvořáčková M, Růžička F, Benešík M, Pantůček R, Dvořáková-Heroldová M (2019) Antimicrobial effect of commercial phage preparation Stafal® on biofilm and planktonic forms of methicillin-resistant Staphylococcus aureus. Folia Microbiol 64(1):121–126. https://doi.org/10.1007/s12223-018-0622-3
Fischetti VA (2018) Development of phage lysins as novel therapeutics: a historical perspective. Viruses 10(6):310. https://doi.org/10.3390/v10060310
Patey O, McCallin S, Mazure H, Liddle M, Smithyman A, Dublanchet A (2019) Clinical indications and compassionate use of phage therapy: personal experience and literature review with a focus on osteoarticular infections. Viruses 11(1):18. https://doi.org/10.3390/v11010018
Wittebole X, De Roock S, Opal SM (2014) A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5(1):226–235. https://doi.org/10.4161/viru.25991
Tan SY, Tatsumura Y (2015) Alexander Fleming (1881–1955): discoverer of penicillin. Singapore Med J 56(7):366–367. https://doi.org/10.11622/smedj.2015105
Lowy FD (2003) Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Investig 111(9):1265–1273. https://doi.org/10.1172/JCI18535
Abdelkader K, Gerstmans H, Saafan A, Dishisha T, Briers Y (2019) The preclinical and clinical progress of bacteriophages and their lytic enzymes: the parts are easier than the whole. Viruses 11(2):96. https://doi.org/10.3390/v11020096
Acknowledgements
We are thankful to Environmental Virology Cell, CSIR-NEERI, Nagpur and Director CSIR-NEERI for providing the research facility and infrastructure. Financial assistance to Akanksha Rai in form of fellowship from the University Grants Commission (UGC), India is gratefully acknowledged. We thank Traci Raley, MS, ELS, for editing a draft of this manuscript.
Funding
Financial assistance as fellowship from the University Grants Commission (UGC), India.
Author information
Authors and Affiliations
Contributions
Conceptualization: Krishna Khairnar and Akanksha Rai; Writing and original draft preparation: Akanksha Rai; Writing, reviewing, and editing: Akanksha Rai and Krishna Khairnar.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Agnes M.S. Figueiredo
Rights and permissions
About this article
Cite this article
Rai, A., Khairnar, K. Overview of the risks of Staphylococcus aureus infections and their control by bacteriophages and bacteriophage-encoded products. Braz J Microbiol 52, 2031–2042 (2021). https://doi.org/10.1007/s42770-021-00566-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00566-4