Abstract
Paracetamol or acetaminophen is a medication commonly used in pain and fever. It is typically used for mild to moderate pain relief. It can produce selective inhibition to the prostaglandin synthesis. We envisage the density functional theory (DFT) with B3LYP/6-31G+(d,p) basis set to optimize the newly modified derivatives. Thermodynamic properties, molecular orbital features, dipole moment, atomic partial charge and electrostatic potential have been calculated in order to compare their physicochemical and biological properties. Molecular docking, nonbonding interactions, and dynamics simulation have been performed against prostaglandin H2 (PGH2) synthase protein 5F19 to investigate their binding affinity, binding modes, and stability of the protein-drug complex. ADMET prediction has been utilized to compare the absorption, metabolism, and carcinogenic properties of new derivatives with parent drug (PCT). From physicochemical data, all modified structures are thermodynamically stable; most of them are chemically more reactive and show better binding affinity than the parent drug. ADMET calculations predict the improved pharmacokinetic properties of modified derivatives. Based on physicochemical, docking, dynamics simulation and ADMET prediction results, this study can be helpful to design a new analgesic and antipyretic drug.
Similar content being viewed by others
1 Introduction
Paracetamol (PCT) or acetaminophen is one of the most commonly used analgesic and antipyretic drugs with relatively little anti-inflammatory activity [1, 2]. The analgesic and antipyretic actions of PCT resemble those of non-steroidal anti-inflammatory drugs (NSAIDs). PCT acts as weak inhibitor of prostaglandin (PG) synthesis by reducing cyclooxygenase (COX-1 and COX-2) [3, 4]. It has some demerits depending on the type and nature of unusual physical condition of the body and on the limit of dose. Overdose and long term dose of PCT produce hepatic and/or renal injury in human and other experimental animals [5,6,7]. N-acetyl-p-benzoquinone-imine (NAPQI) is the toxic intermediary metabolite of PCT that accumulate in the liver due to overdose when glutathione storage is depleted [8]. Considering molecular mechanism of analgesic activity as well as the hepatotoxicity of PCT, there are several modified derivatives are studied to search improved analgesic and antipyretic activities and to reduce side effects [6]. Different electron donating (–CH3, –OCH3, and –SCH3) and electron withdrawing (–Br, –Cl, –F, –I, –CF3 and –CCl3) substituents are placed at X, Y and Z positions of core structure in order to rationalize the analgesic and antipyretic activity.
Drug modification is another alternative way to search better agent, which can increase the selective action and reduce the side effects of drugs. Currently, it is observed that the modification of drugs inserting halogen, alkyl, alkoxy, and hydroxyl groups which play an important role in improving drug performance [9,10,11,12]. In this study, we reported the optimization of PCT and its modified derivatives to investigate their biochemical behaviour on the basis of quantum mechanical calculation. The free energy, enthalpy, dipole moment, electrostatic potential, HOMO–LUMO gap, hardness, softness, and atomic partial charge have been calculated. Molecular docking and nonbonding interactions have been performed to investigate the binding affinity, mode(s) and interactions of drugs with the amino acid residues of receptor protein. The docking prediction was further validated by dynamics simulation. Most of the derivatives showed improved thermal stability, chemical reactivity, binding affinities and interactions. Hydrogen bonding plays crucial role in the biological system which have ultimate effect on thermodynamic and structural stability of a system [13,14,15,16]. From the regarding quantum chemical studies, we are assuming that, some of the designed compounds may have profound effect as drugs.
2 Computational details
2.1 Geometry optimization
In computer aided drug design, quantum mechanical methods has greater attention on calculation of thermodynamic properties, molecular orbital features, dipole moment, atomic partial charge, molecular electrostatic potential properties [17]. Initial geometry of Paracetamol (PCT) was taken from online structure database named ChemSpider [18]. Geometry optimization and further modification of all structures were carried out using Gaussian 09 program [19]. Density functional theory (DFT) with Becke’s (B) [20] three parameter hybrid model Lee, Yang and Parr’s (LYP) correlation functional [21] under Pople’s 6-31G+(d,p) basis set was used for geometry optimization [22]. Initial optimization of all compounds was performed in the gas phase then re-optimized in water phase under polarizable continuum model (PCM) [11, 23] at same level of theory. The difference of electronic energy (∆E), enthalpy (∆H), Gibb’s free energy (∆G) and dipole moment (∆μ) were calculated by subtracting gas phase energy from water phase energy.
Molecular orbitals were calculated at same level of theory. Hardness (η) and softness (S) of all compounds were also calculated from the energies of HOMO (highest occupied molecular orbital) and LUMO (lowest unoccupied molecular orbital) considering Parr and Pearson interpretation [24, 25] of DFT and Koopmans theorem [26] on the correlation of ionization potential (I) and electron affinities (E) with HOMO and LUMO energy (). The following equations are used for the calculation of hardness (η) and softness (S):
2.2 Preparation of target protein
The 3D crystal structure of aspirin acetylated human cyclooxygenase-2 (PDB ID: 5F19) was obtained in pdb format from online protein data bank (PDB) database [27]. The structure was prepared by erasing water molecules, hetero atoms, inactive chain, and adding hydrogen atoms by PyMol (version 1.3) software package [28] and proceed with chain A of 5F19 for further energy minimization with Swiss-PDB Viewer software package (Version 4.1.0) [29].
2.3 Docking, analysis and visualization
Molecular docking is an important tool in computational drug design by which one can predict the predominant binding mode(s) of a ligand with a target protein. The optimized structures were subjected for molecular docking study against human prostaglandin synthase protein 5F19 (chain A) considering the protein as macromolecule and the drug as ligand where the parent drug considered as control and derivatives binding affinity was compared with the parent drug, which is a known Cox-2 inhibitor. The centre grid box was set at 64.84 Å, 73.29 Å and 57.94 Å in x, y and z direction respectively. Flexible docking was performed using PyRx software (version 0.8) [30]. Finally we saved both the proteins and ligand structures in pdb format for further non-bonding interactions and hydrogen bond surface calculation. Accelrys Discovery Studio (version 4.1) software was utilize to analyze and visualize the docking result [31].
2.4 Molecular dynamics (MD) simulations
To validate the predicted molecular docking, MD simulation was performed on the docked PCT-5F19 and D6-5F19 complex using AMBER14 force field implemented in YASARA dynamics program. Water molecules were added and system was neutralized by adding NaCl salt at 0.9% concentration. MD simulation was equilibrated for 100 ps followed by 20 ns production at 298 K with time step of 5 fs.
2.5 ADMET prediction
AdmetSAR online database was utilized to predict absorption, distribution, metabolism, excretion, and toxicity (ADMET) of PCT and its modified derivatives [32].
3 Results and discussion
3.1 Thermodynamic properties
Simple modification of drug can improve the physicochemical and binding properties [33]. With the modification of PCT, significant change observed in structural properties as well as energies, electrostatic potential, partial charge distribution, dipole moment. Free energy is an important criterion to represent the interaction of binding partners where both the sign and magnitudes carry the important information. Negative sign indicate spontaneous binding interactions, where as positive sign opposite to that [34]. Here, all calculated energies were found negative which indicate preferable binding interactions. The results (Table 1) demonstrate that all the derivatives of PCT are chemically stable and few of them are more stable than parent drug. By considering the structure of PCT and its derivatives D1–D9 in the gaseous phase to water solvent it has been observed that there is a gradual decrease of enthalpy, Gibbs free energy, thermal energy. The free energy difference of PCT and D4 are − 8.59 kcal/mol and − 8.84 kcal/mol respectively. Where D4 has the highest free energy (− 9.04 kcal/mol) due to the addition of higly electronegative fluorine atom. Moreover, increasing of negative values from PCT to D9, hence suggesting –F, and -SCH3 substituted PCT molecules (D3, D4, D6 and D9) energetically and configurationally preferable.
Improved dipole moment can enhance the polar nature of molecule, binding affinity and interactions with the amino acid residues of receptor protein [35]. Significant results are observed from gaseous phase to the water solvent. The change of dipole moment of PCT is 0.99 Debye where D5 shows the largest value (2.10 Debye) respectively and most of the modified drugs (D1, D3, D4, D5, D9) show higher value than PCT. Resulting, increased dipole moment causes an increasing of binding affinity against 5F19 protein (Table 2).
3.2 Frontier molecular orbital analysis
The HOMO–LUMO gap is related to the chemical hardness and softness of a molecule [36, 37]. Larger HOMO–LUMO gap related to high kinetic stability and low chemical reactivity where as small HOMO–LUMO gap is important for low chemical stability, addition of electrons to a high-lying LUMO and/or removal of electrons from a low-lying HOMO is energetically favourable in any potential reaction [38, 39]. In this analysis, it is found that, brominated (D7), iodinated (D8) and fluorinated (D6) derivatives show the lower HOMO–LUMO gap and higher softness, which may contribute to their chemical activity and polarizability than PCT.
3.3 Molecular electrostatic potential analysis
Molecular electrostatic potential (MEP) was calculated at B3LYP/6-31G+(d,p) level of theory to forecast the reactive sites for electrophilic and nucleophilic attack of all optimized structures [40]. Red colour represent maximum negative area which favourable site for electrophilic attack, blue colour indicate the maximum positive area which favourable site for nucleophilic attack and green colour represent zero potential area. MEP displays molecular size, shape as well as positive, negative and neutral electrostatic potential regions simultaneously in terms of colour grading. It is seen from MEP map, region having the negative potential are over electronegative atom (oxygen atoms) and having positive potential are over hydrogen atoms. Here, D3 have the maximum negative potentiality (− 0.2313 a.u., deep red) and D6 have the highest positive potentiality (+ 0.2769 a.u., deep blue) which suggesting the maximum possibility for the nucleophilic and electrophilic attack to the respected region (Figs. 1, 2).
3.4 Atomic partial charge
Mulliken and NBO methods have been utilized to compute the atomic partial charges of all structures [41]. Dipole moment and polarizability influenced by the atomic partial charge [42]. In this study (Figure S5), all the hydrogen atoms show positive charge and other electronegative atoms (N,O) exhibit the negative charge in both methods. In all compounds, C-1 and C-10 have the negative charge but in D6, C-1 has the positive charge due to the attachment of highly electronegative fluorine atoms. Among the halogen substituents, fluorine and bromine exhibit negative charge (in D4, D6, D7) but chlorine and iodine exhibit the positive charge in D5, D9, and D8 respectively (Figs. 3, 4).
3.5 Binding affinity and non-bonding interactions
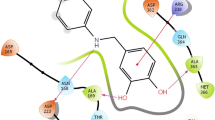
Binding affinities and non-bonding interactions are summarized in Table 3. The addition of halogen not only increase the physicochemical properties but also binding affinity and speciality. The incorporation of –CF3 group increased inhibiting and medicinal properties [43,44,45,46]. Carbon trifluoride group has a great significant applications in the field of agronomical dyes, pigments, pharmaceuticals, polymers and material science when it is incorporated to different organic molecules due to the strong electronegative and hydrophobic characteristics, which can be used in drug design to improve the selective functionality [47,48,49,50]. In our study, all the compounds show several significant hydrogen bonds. Some recent studies reported that, halogen bonding similar to hydrogen bonding plays crucial role for biological and chemical system [51, 52]. The binding affinity of D6, D7, D8, and D9 have considerably increased to − 7.5, − 7.1, − 7.1, − 7.0 and − 7.3 kcal/mol respectively from − 6.4 kcal/mol of PCT. Improved hydrogen bond is observed in D9 and D6 not only contribute in increasing binding affinity but also enhance the binding specificity [53]. All the molecules show multiple nonbonding interactions after the docking with 5F19. Two strong hydrogen bonds with His386 (2.36 Å) and Thr206 (2.84 Å), along with increasing number of halogen bonds and one hydrophobic interaction are found in D6-5F19 complex. Thus strong hydrogen bonding and halogen bonding have important role in increasing binding affinity of D6 with 5F19. The D7-5F19 complex is stabilize by two hydrogen bonds and one hydrophobic interaction (Table 3). The most important fact is that the -F atoms of –CF3 group in D6-5F19 complex interact with amino acid to form strong halogen bond interactions. We also observed six fluorine bond (halogen bond) in D6-5F19 and one in D4-5F19 complex (Fig. 5) which may have positive effect on protein-ligands stability, as well as in the binding affinity and selectivity. D6 has higher softness than PCT which may promote the polarizable nature of the drug that can stimulate greater nonbonding interactions with the receptor. Nonbonding interactions of (PCT, D6, D7, D8, and D9) with 5F19 involved the following amino acid residues Ala199, Thr206, His386, His388, Tyr385, Leu531, Leu534, Gly533 and Met522 (Table 3). Hydrogen bond surface having residues such as Ala202, Val349, Gin461, Gly135, Pro156, His39 and His386 help in creating strong donor regions where as residues such as Ala199, Thr206, Tyr385, Met522, Gly45, Gln461, Pro154, Phe529, Gly526, Asn382, His207, His388 and Tyr385 help in creating strong acceptor regions on the drug protein interaction surface (Fig. 6). This observation helped to confirm that PCT and its selected derivatives (D6, D8 and D9) are binding at the desired binding site of receptor protein after molecular docking. Ala202, Ala203, Ala528His388, Leu353 and Val524 are found to donate their π-electrons cloud toward the alkyl chain and the carbon attached.
In case of D1, D2, D3, D4 and D5, X and Y positions of PCT are substituted by following functional groups –CH3, –OCH3, –SCH3, –F, –Cl and they their binding affinity with 5F19 are − 6.9, − 6.6, − 5.7, − 7.0, − 6.8 kcal/mol respectively. Three strong hydrogen bonds are found in D2 with Gly45 (2.00Å), Gln461 (2.04 Å), Gln461 (1.97 Å) and one hydrophobic interaction with Leu153. Where, D1, D3 and D4 also show strong hydrogen bond Tyr385 (2.25 Å), Gly135 (2.28 Å) and D5 and Ala199 (2.27 Å). D5 shows the maximum number of hydrophobic interactions with two amide-pi stacked (Ala527 and Gly526). All the compounds show non-conventional hydrogen bond except D1, D5 and D9. Generally, CH···O bonding term as non-conventional hydrogen bond, slightly weaker than its classical OH···O hydrogen bond [54, 55]. Those interactions may be the factors of improved binding affinity of D5 with 5F19 protein.
3.6 Molecular dynamics calculation
Parent compound (PCT) along with the best compound from docking analysis (D6) are analysed furthermore for conformational and binding stability through molecular dynamics study. 20 ns MD simulation was performed for PCT-5F19 and D6-5F19 complex. Root-mean-square deviation (RMSD) values of α-carbon and backbone atoms are investigated (Fig. 7). The RMSD values of PCT-5F19 and D6-5F19 complexes are plotted as a time-dependent function of the MD simulation. Very little RMSD deviation between alpha-carbon and backbone carbon atoms throughout the whole simulation process was evident and it indicates that the protein structure remains stable in both drug–protein complexes. The maximum RMSD value for the PCT-5F19 complex is 2.3 Å at around 16 ns whereas for the D6-5F19 complex the maximum RMSD value is same but at around 4.8 ns and 19.8 ns. Evaluation of molecular docking can also be performed through molecular dynamics simulations. In this study, both the drugs remain in the binding pocket after MD simulation similar to the docked pose of PCT-5F19 and D6-5F19 complexes (Fig. 8).
3.7 ADMET analysis
Selected pharmacokinetics parameters are reported in Table 4. From ADMET results, it is found that all the structures show positive response for blood brain barrier (BBB) criteria, predicting that drugs can pass through the BBB. All the compounds are non-carcinogenic and show III category acute oral toxicity so, the modified drugs expected to be safe for topical biological use. All drugs are P-glycoprotein non-inhibitor and show positive response for human intestinal absorption. P-glycoprotein inhibition can block the absorption, permeability and retention of the drugs [56]. However, PCT and its modified derivatives show weak inhibitory property for human ether-a-go-go-related gene (hERG). Inhibition of hERG can lead to long QT syndrome [57], so more study of this aspect is necessary. The rat acute toxicity level of all drugs is higher than the PCT. So, modified derivatives have higher median lethal dose (LD50) than PCT.
4 Conclusion
In this investigation, PCT and its modified derivatives are studied to explore their physicochemical properties and binding affinities with receptor protein. All the compounds are thermally stable and most of them have lower HOMO–LUMO gap with higher softness values. All the derivatives (except D3) have better binding affinity and interactions with the receptor protein compared to parent drug. All the derivatives have improved pharmacokinetic properties and safe for biological use. Moreover, D4, D7, D8, and D9 also show higher binding affinity and important non-bonding interactions than PCT. Where, D6-5F19 complex shows the highest binding affinity than others and remain inside the binding pocket of target protein after dynamics simulation. From the above results, it is observed that the insertion of halogen improved the physicochemical and binding properties. Considering the above investigation, D4, D6, D8, and D9 can be potential candidates for better performance.
References
Toes MJ, Jones AL, Prescott L (2005) Drug interactions with paracetamol. Am J Ther 12:56–66
Yoosefian M, Ansarinik Z, Etminan N (2016) Density functional theory computational study on solvent effect, molecular conformations, energies and intramolecular hydrogen bond strength in different possible nano-conformers of acetaminophen. J Mol Liq 213:115–121
Hinz B, Cheremina O, Brune K (2008) Acetaminophen (paracetamol) is a selective cyclooxygenase-2 inhibitor in man. FASEB J 22:383–390. https://doi.org/10.1096/fj.07-8506com
Graham GG, Scott KF (2005) Mechanism of action of paracetamol. Am J Ther 12:46–55
Ferencz L, Muntean DL (2015) Possible substitutes for paracetamol: the results of a comprehensive screening based on structural similarity and docking simulation on the surface of enzymes. Farmacia 63:422–428
Diniz JEM, Borges RS, Alves CN (2004) A DFT study for paracetamol and 3,5-disubstituted analogues. J Mol Struct Theochem 673:93–97. https://doi.org/10.1016/j.theochem.2003.12.002
Thomas SHL (1993) Paracetamol (acetaminophen) poisoning. Pharmacol Ther 60:91–120. https://doi.org/10.1016/0163-7258(93)90023-7
Wong A, Landersdorfer C, Graudins A (2017) Pharmacokinetic modelling of modified acetylcysteine infusion regimens used in the treatment of paracetamol poisoning. Eur J Clin Pharmacol 73(9):1103–1110
Juillerat-Jeanneret L, Schmitt F (2007) Chemical modification of therapeutic drugs or drug vector systems to achieve targeted therapy: looking for the grail. Med Res Rev 27:574–590
Rahman A, Hoque MM, Khan MAK et al (2016) Non-covalent interactions involving halogenated derivatives of capecitabine and thymidylate synthase: a computational approach. Springerplus 5:146
Uzzaman M, Uddin MN (2019) Optimization of structures, biochemical properties of ketorolac and its degradation products based on computational studies. DARU J Pharm Sci 27(1):71–82. https://doi.org/10.1007/s40199-019-00243-w
Li X-Q, Andersson TB, Ahlström M, Weidolf L (2004) Comparison of inhibitory effects of the proton pump-inhibiting drugs omeprazole, esomeprazole, lansoprazole, pantoprazole, and rabeprazole on human cytochrome P450 activities. Drug Metab Dispos 32:821–827
Raissi H, Yoosefian M, Mollania F, Farzad F (2012) The effect of substitution on structure, intramolecular hydrogen bonding strength, electron density and resonance in 3-amino 2-iminomethyl acryl aldehyde. J Theor Comput Chem 11:925–939. https://doi.org/10.1142/S0219633612500629
Raissi H, Yoosefian M, Mollania F (2012) Hydrogen bond studies in substituted imino-acetaldehyde oxime. Comput Theor Chem 996:68–75. https://doi.org/10.1016/j.comptc.2012.07.017
Yoosefian M, Raissi H, Davamdar E et al (2012) Synthesis and theoretical study of intramolecular hydrogen bond at two possible positions in pyrazolo[1,2-b]phthalazine. Chin J Chem 30:779–784. https://doi.org/10.1002/cjoc.201100036
Koll A, Karpfen A, Wolschann P (2006) Structural and energetic consequences of the formation of intramolecular hydrogen bonds. J Mol Struct 790:55–64
Gleeson MP, Gleeson D (2009) QM/MM calculations in drug discovery: a useful method for studying binding phenomena? J Chem Inf Model 49:670–677. https://doi.org/10.1021/ci800419j
Pence HE, Williams A (2010) ChemSpider: an online chemical information resource. J Chem Educ 87:1123–1124. https://doi.org/10.1021/ed100697w
Frisch A (2009) gaussian 09 W Reference. Wallingford, USA, 25p
Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A 38:3098–3100. https://doi.org/10.1103/PhysRevA.38.3098
Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37:785–789. https://doi.org/10.1103/PhysRevB.37.785
Kruse H, Goerigk L, Grimme S (2012) Why the standard B3LYP/6-31G* model chemistry should not be used in DFT calculations of molecular thermochemistry: understanding and correcting the problem. J Org Chem 77:10824–10834. https://doi.org/10.1021/jo302156p
Tomasi J, Mennucci B, Cammi R (2005) Quantum mechanical continuum solvation models. Chem Rev 105:2999–3094
Calais J-L (1993) Density-functional theory of atoms and molecules. R.G. Parr and W. Yang, Oxford University Press, New York, Oxford, 1989. IX + 333 pp. Price £45.00. Int J Quantum Chem 47:101. https://doi.org/10.1002/qua.560470107
Pearson RG (1995) The HSAB principle—more quantitative aspects. Inorganica Chim Acta 240:93–98. https://doi.org/10.1016/0020-1693(95)04648-8
Pearson RG (1986) Absolute electronegativity and hardness correlated with molecular orbital theory. Proc Natl Acad Sci 83:8440–8441
Lucido MJ, Orlando BJ, Vecchio AJ, Malkowski MG (2016) Crystal structure of aspirin-acetylated human cyclooxygenase-2: insight into the formation of products with reversed stereochemistry. Biochemistry 55:1226–1238. https://doi.org/10.1021/acs.biochem.5b01378
Delano WL (2002) The PyMOL molecular graphics system. Version 1. Schrödinger LLC. http://www.pymol.org. doi: citeulikearticle-id:240061
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. https://doi.org/10.1002/elps.1150181505
Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. In: Hempel JE, Williams CH, Hong CC (eds) Chemical biology: methods and protocols. Springer, New York, pp 243–250
Version ADS (2017) 4.0, Accelrys, San Diego, USA
Cheng F, Li W, Zhou Y et al (2012) admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 52:3099–3105. https://doi.org/10.1021/ci300367a
Uzzaman M, Hoque MJ (2018) Physiochemical, molecular docking, and pharmacokinetic studies of Naproxen and its modified derivatives based on DFT. Int J Sci Res Manag. https://doi.org/10.18535/ijsrm/v6i9.c01
Garbett NC, Chaires JB (2012) Thermodynamic studies for drug design and screening. Expert Opin Drug Discov 7:299–314. https://doi.org/10.1517/17460441.2012.666235
Lien EJ, Guo Z-R, Li R-L, Su C-T (1982) Use of dipole moment as a parameter in drug–receptor interaction and quantitative structure-activity relationship studies. J Pharm Sci 71:641–655. https://doi.org/10.1002/jps.2600710611
Ayers PW, Parr RG, Pearson RG (2006) Elucidating the hard/soft acid/base principle: a perspective based on half-reactions. J Chem Phys 124:194107. https://doi.org/10.1063/1.2196882
Parr RG, Zhou Z (1993) Absolute hardness: unifying concept for identifying shells and subshells in nuclei, atoms, molecules, and metallic clusters. Acc Chem Res 26:256–258. https://doi.org/10.1021/ar00029a005
Aihara J (1999) Reduced HOMO–LUMO gap as an index of kinetic stability for polycyclic aromatic hydrocarbons. J Phys Chem A 103:7487–7495. https://doi.org/10.1021/jp990092i
Aihara J (2000) Correlation found between the HOMO–LUMO energy separation and the chemical reactivity at the most reactive site for isolated-pentagon isomers of fullerenes. Phys Chem Chem Phys 2:3121–3125. https://doi.org/10.1039/B002601H
Cox SR, Williams DE (1981) Representation of the molecular electrostatic potential by a net atomic charge model. J Comput Chem 2:304–323. https://doi.org/10.1002/jcc.540020312
Gross KC, Seybold PG, Hadad CM (2002) Comparison of different atomic charge schemes for predicting pKa variations in substituted anilines and phenols*. Int J Quantum Chem 90:445–458. https://doi.org/10.1002/qua.10108
Mulliken RS (1955) Electronic population analysis on LCAO–MO molecular wave functions I. J Chem Phys 23:1833–1840
Abu Saleh M, Solayman M, Hoque MM, et al (2016) Inhibition of DNA topoisomerase type IIα (TOP2A) by mitoxantrone and its halogenated derivatives: a combined density functional and molecular docking study. Biomed Res Int 2016:6817502. https://doi.org/10.1155/2016/6817502
Shawon J, Khan AM, Rahman A et al (2016) Molecular recognition of azelaic acid and related molecules with DNA polymerase I investigated by molecular modeling calculations. Interdiscip Sci Comput Life Sci 10(3):525–537
Nair HK, Quinn DM (1993) m-Alkyl, α, α, α-trifluoroacetophenones: a new class of potent transition state analog inhibitors of acetylcholinesterase. Bioorg Med Chem Lett 3:2619–2622. https://doi.org/10.1016/S0960-894X(01)80727-7
Plano D, Karelia DN, Pandey MK et al (2016) Design, synthesis, and biological evaluation of novel selenium (Se-NSAID) molecules as anticancer agents. J Med Chem 59:1946–1959
McClinton MA, McClinton DA (1992) Trifluoromethylations and related reactions in organic chemistry. Tetrahedron 48:6555–6666. https://doi.org/10.1016/S0040-4020(01)80011-9
Ji Y, Brueckl T, Baxter RD et al (2011) Innate CH trifluoromethylation of heterocycles. Proc Natl Acad Sci 108:14411–14415
Furuya T, Kamlet AS, Ritter T (2011) Catalysis for fluorination and trifluoromethylation. Nature 473:470–477
Lishchynskyi A, Novikov MA, Martin E et al (2013) Trifluoromethylation of aryl and heteroaryl halides with fluoroform-derived CuCF3: scope, limitations, and mechanistic features. J Org Chem 78:11126–11146. https://doi.org/10.1021/jo401423h
Sarwar MG, Ajami D, Theodorakopoulos G et al (2013) Amplified halogen bonding in a small space. J Am Chem Soc 135:13672–13675. https://doi.org/10.1021/ja407815t
Sarwar MG, Dragisic B, Salsberg LJ et al (2010) Thermodynamics of halogen bonding in solution: substituent, structural, and solvent effects. J Am Chem Soc 132:1646–1653. https://doi.org/10.1021/ja9086352
Bissantz C, Kuhn B, Stahl M (2010) A medicinal chemist’s guide to molecular interactions. J Med Chem 53:5061–5084. https://doi.org/10.1021/jm100112j
Kuduva SS, Craig DC, Nangia A, Desiraju GR (1999) Cubanecarboxylic acids. Crystal engineering considerations and the role of C–H···O hydrogen bonds in determining O–H···O networks. J Am Chem Soc 121:1936–1944. https://doi.org/10.1021/ja981967u
Meadows ES, De Wall SL, Barbour LJ et al (2000) Structural and dynamic evidence for C–H···O hydrogen bonding in lariat ethers: implications for protein structure. J Am Chem Soc 122:3325–3335. https://doi.org/10.1021/ja9940672
Amin ML (2013) P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights 7:27
Sanguinetti MC, Tristani-Firouzi M (2006) hERG potassium channels and cardiac arrhythmia. Nature 440:463–469
Acknowledgements
We are thankful to Dr. Mohammad Abdul Halim (CEO, The Red-Green Research Centre, Dhaka, Bangladesh) for his valuable suggestions and help in MD simulation.
Author information
Authors and Affiliations
Contributions
Monir Uzzaman (MZ) designed the project, performed all quantum calculation, data collection and manuscript writing. Molecular dynamics simulation was performed by Jakaria Shawon (JS). Zainul Abedin Siddique (ZAS) has reviewed the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Uzzaman, M., Shawon, J. & Siddique, Z.A. Molecular docking, dynamics simulation and ADMET prediction of Acetaminophen and its modified derivatives based on quantum calculations. SN Appl. Sci. 1, 1437 (2019). https://doi.org/10.1007/s42452-019-1442-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-019-1442-z