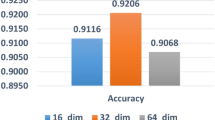
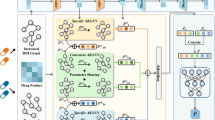
Abstract
Drug–Drug interaction (DDI) prediction is essential in pharmaceutical research and clinical application. Existing computational methods mainly extract data from multiple resources and treat it as binary classification. However, this cannot unambiguously tell the boundary between positive and negative samples owing to the incompleteness and uncertainty of derived data. A granular computing method called three-way decision is proved to be effective in making uncertain decision, but it relies on supplementary information to make delay decision. Recently, biomedical knowledge graph has been regarded as an important source to obtain abundant supplementary information about drugs. This paper proposes a three-way decision-based method called 3WDDI, in combination with knowledge graph embedding as supplementary features to enhance DDI prediction. The drug pairs are divided into positive, negative and boundary regions by Convolutional Neural Network (CNN) according to drug chemical structure feature. Further, delay decision is made for objects in the boundary region by integrating knowledge graph embedding feature to promote the accuracy of decision-making. The empirical results show that 3WDDI yields up to 0.8922, 0.9614, 0.9582, 0.8930 for Accuracy, AUPR, AUC and F1-score, respectively, and outperforms several baseline models.


Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Data availability
Data sharing not applicable to this article as no datasets were generated.
References
Abdelaziz I, Fokoue A, Hassanzadeh O et al (2017) Large-scale structural and textual similarity-based mining of knowledge graph to predict drug-drug interactions. J Web Semant 44:104–117
Bordes A, Usunier N, García-Durán A et al (2013) Translating embeddings for modeling multi-relational data. Adv Neural Inf Process Syst 26:2787–2795
Callahan A, Cruz-Toledo J, Ansell P et al (2013) Bio2rdf release 2: improved coverage, interoperability and provenance of life science linked data. Extended semantic web conference. Springer, New York, pp 200–212
Chen Q, Lai D, Lan W et al (2019) ILDMSF: inferring associations between long non-coding rna and disease based on multi-similarity fusion. IEEE/ACM Trans Comput Biol Bioinform 18(3):1106–1112
Chen Q, Qiao Y, Hu F et al (2020) Community detection in complex network based on APT method. Pattern Recogn Lett 138:193–200
Cheng F, Zhao Z (2014) Machine learning-based prediction of drug-drug interactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties. J Am Med Inf Assoc 21(e2):e278–e286
Dai Y, Guo C, Guo W et al (2020) Drug-drug interaction prediction with Wasserstein adversarial autoencoder-based knowledge graph embeddings. Brief Bioinform 22(4):bbaa256
Deng Y, Xu X, Qiu Y et al (2020) A multimodal deep learning framework for predicting drug-drug interaction events. Bioinformatics 36(15):4316–4322
Ferdousi R, Safdari R, Omidi Y (2017) Computational prediction of drug-drug interactions based on drugs functional similarities. J Biomed Inform 70:54–64
Gottlieb A, Stein GY, Oron Y et al (2012) INDI: a computational framework for inferring drug interactions and their associated recommendations. Mol Syst Biol 8(1):592
Huaxiong L, Libo Z, Bing H et al (2016) Sequential three-way decision and granulation for cost-sensitive face recognition. Knowl-Based Syst 91:241–251
Ioannidis VN, Song X, Manchanda S et al. (2020) DRKG- drug repurposing knowledge graph for covid-19. https://github.com/gnn4dr/DRKG/
Ji S, Pan S, Cambria E et al (2022) A survey on knowledge graphs: representation, acquisition, and applications. IEEE Trans Neural Netw Learn Syst 33(2):494–514
Kanehisa M, Sato Y, Kawashima M et al (2015) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44(D1):D457–D462
Karim MR, Cochez M, Jares JB et al. (2019) Drug-drug interaction prediction based on knowledge graph embeddings and convolutional-lstm network. In: International conference on bioinformatics, computational biology and health informatics, pp 113–123
Kastrin A, Ferk P, Leskošek B (2018) Predicting potential drug-drug interactions on topological and semantic similarity features using statistical learning. PLoS One 13(5):1–23
Lan W, Lai D, Chen Q et al (2020) LDICDL: Lncrna-disease association identification based on collaborative deep learning. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2020.3034910
Lan W, Dong Y, Chen Q et al (2021a) IGNSCDA: predicting circrna-disease associations based on improved graph convolutional network and negative sampling. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2021.3111607
Lan W, Dong Y, Chen Q et al (2021b) KGANCDA: predicting circRNA-disease associations based on knowledge graph attention network. Brief Bioinform 23(1):bbab494
Lan W, Wu X, Chen Q et al (2022) GANLDA: Graph attention network for lncrna-disease associations prediction. Neurocomputing 469:384–393
Law V, Knox C, Djoumbou Y et al (2013) Drugbank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42(D1):D1091–D1097
Lee G, Park C, Ahn J (2019) Novel deep learning model for more accurate prediction of drug-drug interaction effects. BMC Bioinform 20(1):1–8
Li Z, Huang D (2020) A three-way decision method in a fuzzy condition decision information system and its application in credit card evaluation. Granul Comput 5(4):513–526
Li Q, Cheng T, Wang Y et al (2010) Pubchem as a public resource for drug discovery. Drug Discov Today 15(23):1052–1057
Li Y, Zhang L, Xu Y et al (2017) Enhancing binary classification by modeling uncertain boundary in three-way decisions. IEEE Trans Knowl Data Eng 29(7):1438–1451
Lin Y, Liu Z, Sun M et al. (2015) Learning entity and relation embeddings for knowledge graph completion. In: Twenty-ninth AAAI conference on artificial intelligence, pp 2181–2187
Rohani N, Eslahchi C (2019) Drug-drug interaction predicting by neural network using integrated similarity. Sci Rep 9(1):1–11
Ryu JY, Kim HU, Lee SY (2018) Deep learning improves prediction of drug–drug and drug–food interactions. Proc Natl Acad Sci USA 115(18):E4304–E4311
Trouillon T, Welbl J, Riedel S et al. (2016) Complex embeddings for simple link prediction. In: International conference on machine learning, pp 2071–2080
Wang Z, Zhang J, Feng J, et al. (2014) Knowledge graph embedding by translating on hyperplanes. In: Twenty-Eighth AAAI conference on artificial intelligence, pp 1112–1119
Yang B, Yih W, He X, et al. (2014) Embedding entities and relations for learning and inference in knowledge bases. arXiv preprint arXiv:1412.6575
Yao Y (2010) Three-way decisions with probabilistic rough sets. Inf Sci 180(3):341–353
Yao Y (2021) Set-theoretic models of three-way decision. Granul Comput 6(1):133–148
Yu H, Wang X, Wang G et al (2020) An active three-way clustering method via low-rank matrices for multi-view data. Inf Sci 507:823–839
Zhang P, Wang F, Hu J et al (2015) Label propagation prediction of drug-drug interactions based on clinical side effects. Sci Rep 5(1):1–10
Zhang HR, Min F, Shi B (2017a) Regression-based three-way recommendation. Inf Sci 378:444–461
Zhang W, Chen Y, Liu F et al (2017b) Predicting potential drug-drug interactions by integrating chemical, biological, phenotypic and network data. BMC Bioinform 18(1):1–12
Zhang Y, Zhang Z, Miao D et al (2019) Three-way enhanced convolutional neural networks for sentence-level sentiment classification. Inf Sci 477:55–64
Zhang Y, Qiu Y, Cui Y et al (2020) Predicting drug-drug interactions using multi-modal deep auto-encoders based network embedding and positive-unlabeled learning. Methods 179:37–46
Zhang C, Gao R, Qin H et al (2021) Three-way clustering method for incomplete information system based on set-pair analysis. Granul Comput 6(2):389–398
Zhou B, Yao Y, Luo J (2014) Cost-sensitive three-way email spam filtering. J Intell Inf Syst 42(1):19–45
Acknowledgements
The work reported in this paper was partially supported by a National Natural Science Foundation of China project 61963004 and 62072124, a key project of Natural Science Foundation of Guangxi 2017GXNSFDA198033, and a key research and development plan of Guangxi AB17195055.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hao, X., Chen, Q., Pan, H. et al. Enhancing drug–drug interaction prediction by three-way decision and knowledge graph embedding. Granul. Comput. 8, 67–76 (2023). https://doi.org/10.1007/s41066-022-00315-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41066-022-00315-4