Abstract
Purpose of Review
To summarize recent literature relating early-life environmental exposures on DNA methylation in the placenta, to identify how variation in placental methylation is regulated in an exposure-specific manner, and to encourage additional work in this area.
Recent Findings
Multiple studies have evaluated associations between prenatal environmental exposures and placental methylation in both gene-specific and epigenome-wide frameworks. Specific exposures lead to unique variability in methylation, and cross-exposure assessments have uncovered certain genes that demonstrate consistency in differential placental methylation. Exposure studies that assess methylation effects in a trimester-specific approach tend to find larger effects during the 1st trimester exposure. Earlier studies have more targeted gene-specific approaches to methylation, while later studies have shifted towards epigenome-wide, array-based approaches. Studies focusing on exposures such as air pollution, maternal smoking, environmental contaminants, and trace metals appear to be more abundant, while studies of socioeconomic adversity and circadian disruption are scarce but demonstrate remarkable effects.
Summary
Understanding the impacts of early-life environmental exposures on placental methylation is critical to establishing the link between the maternal environment, epigenetic variation, and long-term health. Future studies into this field should incorporate repeated measures of exposure throughout pregnancy, in order to determine the critical windows in which placental methylation is most heavily affected. Additionally, the use of methylation-based scores and sequencing technology could provide important insights into epigenetic gestational age and uncovering more genomic regions where methylation is affected. Studies examining the impact of other exposures on methylation, including pesticides, alcohol, and other chemicals are also warranted.



Similar content being viewed by others
References
Newell-Price J, Clark AJ, King P. DNA methylation and silencing of gene expression. Trends Endocrinol Metab. 2000;11(4):142–8.
Hannon E, Knox O, Sugden K, Burrage J, Wong CCY, Belsky DW, et al. Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genet. 2018;14(8):e1007544.
Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21.
Robinson WP, Price EM. The human placental methylome. Cold Spring Harb Perspect Med. 2015;5(5):a023044.
Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–66.
Vryer R, Saffery R. What’s in a name? Context-dependent significance of ‘global’ methylation measures in human health and disease. Clin Epigenetics. 2017;9:2.
Stoccoro A, Coppede F. Mitochondrial DNA methylation and human diseases. Int J Mol Sci. 2021;22(9).
Dong Z, Pu L, Cui H. Mitoepigenetics and its emerging roles in cancer. Front Cell Dev Biol. 2020;8:4.
Baccarelli AA, Byun HM. Platelet mitochondrial DNA methylation: a potential new marker of cardiovascular disease. Clin Epigenetics. 2015;7:44.
Tsaprouni LG, Yang TP, Bell J, Dick KJ, Kanoni S, Nisbet J, et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9(10):1382–96.
Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98(4):680–96.
Elliott HR, Tillin T, McArdle WL, Ho K, Duggirala A, Frayling TM, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics. 2014;6(1):4.
Gruzieva O, Xu CJ, Breton CV, Annesi-Maesano I, Anto JM, Auffray C, et al. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ Health Perspect. 2017;125(1):104–10.
Maghbooli Z, Hossein-Nezhad A, Adabi E, Asadollah-Pour E, Sadeghi M, Mohammad-Nabi S, et al. Air pollution during pregnancy and placental adaptation in the levels of global DNA methylation. PLoS ONE. 2018;13(7):e0199772.
Mostafavi N, Vlaanderen J, Portengen L, Chadeau-Hyam M, Modig L, Palli D, et al. Associations between genome-wide gene expression and ambient nitrogen oxides. Epidemiology. 2017;28(3):320–8.
Goodrich JM, Basu N, Franzblau A, Dolinoy DC. Mercury biomarkers and DNA methylation among Michigan dental professionals. Environ Mol Mutagen. 2013;54(3):195–203.
Tajuddin SM, Amaral AF, Fernandez AF, Rodriguez-Rodero S, Rodriguez RM, Moore LE, et al. Genetic and non-genetic predictors of LINE-1 methylation in leukocyte DNA. Environ Health Perspect. 2013;121(6):650–6.
Tellez-Plaza M, Tang WY, Shang Y, Umans JG, Francesconi KA, Goessler W, et al. Association of global DNA methylation and global DNA hydroxymethylation with metals and other exposures in human blood DNA samples. Environ Health Perspect. 2014;122(9):946–54.
Hanna CW, Bloom MS, Robinson WP, Kim D, Parsons PJ, vom Saal FS, et al. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum Reprod. 2012;27(5):1401–10.
Itoh H, Iwasaki M, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, et al. Association between serum organochlorines and global methylation level of leukocyte DNA among Japanese women: a cross-sectional study. Sci Total Environ. 2014;490:603–9.
Watkins DJ, Wellenius GA, Butler RA, Bartell SM, Fletcher T, Kelsey KT. Associations between serum perfluoroalkyl acids and LINE-1 DNA methylation. Environ Int. 2014;63:71–6.
Clark NA, Demers PA, Karr CJ, Koehoorn M, Lencar C, Tamburic L, et al. Effect of early life exposure to air pollution on development of childhood asthma. Environ Health Perspect. 2010;118(2):284–90.
Lindoso L, Mondal K, Venkateswaran S, Somineni HK, Ballengee C, Walters TD, et al. The effect of early-life environmental exposures on disease phenotype and clinical course of Crohn’s disease in children. Am J Gastroenterol. 2018;113(10):1524–9.
Pesce G, Sese L, Calciano L, Travert B, Dessimond B, Maesano CN, et al. Foetal exposure to heavy metals and risk of atopic diseases in early childhood. Pediatr Allergy Immunol. 2021;32(2):242–50.
Lin X, Teh AL, Chen L, Lim IY, Tan PF, MacIsaac JL, et al. Choice of surrogate tissue influences neonatal EWAS findings. BMC Med. 2017;15(1):211.
Lura MP, Gorlanova O, Muller L, Proietti E, Vienneau D, Reppucci D, et al. Response of cord blood cells to environmental, hereditary and perinatal factors: a prospective birth cohort study. PLoS ONE. 2018;13(7):e0200236.
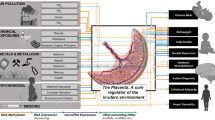
Fisher SJ. The placenta dilemma. Semin Reprod Med. 2000;18(3):321–6.
Maltepe E, Fisher SJ. Placenta: the forgotten organ. Annu Rev Cell Dev Biol. 2015;31:523–52.
Nugent BM, Bale TL. The omniscient placenta: metabolic and epigenetic regulation of fetal programming. Front Neuroendocrinol. 2015;39:28–37.
Barker DJ, Thornburg KL. Placental programming of chronic diseases, cancer and lifespan: a review. Placenta. 2013;34(10):841–5.
Kusuyama J, Alves-Wagner AB, Makarewicz NS, Goodyear LJ. Effects of maternal and paternal exercise on offspring metabolism. Nat Metab. 2020;2(9):858–72.
Mitro SD, Johnson T, Zota AR. Cumulative chemical exposures during pregnancy and early development. Curr Environ Health Rep. 2015;2(4):367–78.
Robinson JF, Hamilton EG, Lam J, Chen H, Woodruff TJ. Differences in cytochrome p450 enzyme expression and activity in fetal and adult tissues. Placenta. 2020;100:35–44.
Sachdeva P, Patel BG, Patel BK. Drug use in pregnancy; a point to ponder! Indian J Pharm Sci. 2009;71(1):1–7.
Myatt L. Placental adaptive responses and fetal programming. J Physiol. 2006;572(Pt 1):25–30.
Saenen ND, Martens DS, Neven KY, Alfano R, Bove H, Janssen BG, et al. Air pollution-induced placental alterations: an interplay of oxidative stress, epigenetics, and the aging phenotype? Clin Epigenetics. 2019;11(1):124.
Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602.
Janssen BG, Godderis L, Pieters N, Poels K, Kicinski M, Cuypers A, et al. Placental DNA hypomethylation in association with particulate air pollution in early life. Part Fibre Toxicol. 2013;10:22.
Sagawa N, Yura S, Itoh H, Kakui K, Takemura M, Nuamah MA, et al. Possible role of placental leptin in pregnancy: a review. Endocrine. 2002;19(1):65–71.
Saenen ND, Vrijens K, Janssen BG, Roels HA, Neven KY, Vanden Berghe W, et al. Lower placental leptin promoter methylation in association with fine particulate matter air pollution during pregnancy and placental nitrosative stress at birth in the ENVIRONAGE cohort. Environ Health Perspect. 2017;125(2):262–8.
Nawrot TS, Saenen ND, Schenk J, Janssen BG, Motta V, Tarantini L, et al. Placental circadian pathway methylation and in utero exposure to fine particle air pollution. Environ Int. 2018;114:231–41.
Neven KY, Saenen ND, Tarantini L, Janssen BG, Lefebvre W, Vanpoucke C, et al. Placental promoter methylation of DNA repair genes and prenatal exposure to particulate air pollution: an ENVIRONAGE cohort study. Lancet Planet Health. 2018;2(4):e174–83.
Zhao Y, Wang P, Zhou Y, Xia B, Zhu Q, Ge W, et al. Prenatal fine particulate matter exposure, placental DNA methylation changes, and fetal growth. Environ Int. 2021;147:106313.
White V, Jawerbaum A, Mazzucco MB, Gauster M, Desoye G, Hiden U. IGF2 stimulates fetal growth in a sex- and organ-dependent manner. Pediatr Res. 2018;83(1–1):183–9.
Garcia-Perez C, Roy SS, Naghdi S, Lin X, Davies E, Hajnoczky G. Bid-induced mitochondrial membrane permeabilization waves propagated by local reactive oxygen species (ROS) signaling. Proc Natl Acad Sci U S A. 2012;109(12):4497–502.
Wang P, Lindsay J, Owens TW, Mularczyk EJ, Warwood S, Foster F, et al. Phosphorylation of the proapoptotic BH3-only protein bid primes mitochondria for apoptosis during mitotic arrest. Cell Rep. 2014;7(3):661–71.
Yang SI, Lee SH, Lee SY, Kim HC, Kim HB, Kim JH, et al. Prenatal PM2.5 exposure and vitamin D-associated early persistent atopic dermatitis via placental methylation. Ann Allergy Asthma Immunol. 2020;125(6):665-731 e1.
Hao N, Whitelaw ML. The emerging roles of AhR in physiology and immunity. Biochem Pharmacol. 2013;86(5):561–70.
Abraham E, Rousseaux S, Agier L, Giorgis-Allemand L, Tost J, Galineau J, et al. Pregnancy exposure to atmospheric pollution and meteorological conditions and placental DNA methylation. Environ Int. 2018;118:334–47.
Ding H, Dai Y, Lei Y, Wang Z, Liu D, Li R, et al. Upregulation of CD81 in trophoblasts induces an imbalance of Treg/Th17 cells by promoting IL-6 expression in preeclampsia. Cell Mol Immunol. 2019;16(1):302–12.
Novakovic B, Evain-Brion D, Murthi P, Fournier T, Saffery R. Variable DAXX gene methylation is a common feature of placental trophoblast differentiation, preeclampsia, and response to hypoxia. FASEB J. 2017;31(6):2380–92.
Roberts VH, Webster RP, Brockman DE, Pitzer BA, Myatt L. Post-translational modifications of the P2X(4) purinergic receptor subtype in the human placenta are altered in preeclampsia. Placenta. 2007;28(4):270–7.
Zhao WX, Huang TT, Jiang M, Feng R, Lin JH. Expression of notch family proteins in placentas from patients with early-onset severe preeclampsia. Reprod Sci. 2014;21(6):716–23.
Ladd-Acosta C, Feinberg JI, Brown SC, Lurmann FW, Croen LA, Hertz-Picciotto I, et al. Epigenetic marks of prenatal air pollution exposure found in multiple tissues relevant for child health. Environ Int. 2019;126:363–76.
Johannesson S, Andersson EM, Stockfelt L, Barregard L, Sallsten G. Urban air pollution and effects on biomarkers of systemic inflammation and coagulation: a panel study in healthy adults. Inhal Toxicol. 2014;26(2):84–94.
Ruckerl R, Hampel R, Breitner S, Cyrys J, Kraus U, Carter J, et al. Associations between ambient air pollution and blood markers of inflammation and coagulation/fibrinolysis in susceptible populations. Environ Int. 2014;70:32–49.
Strauss WJ, Ryan L, Morara M, Iroz-Elardo N, Davis M, Cupp M, et al. Improving cost-effectiveness of epidemiological studies via designed missingness strategies. Stat Med. 2010;29(13):1377–87.
Janssen BG, Gyselaers W, Byun HM, Roels HA, Cuypers A, Baccarelli AA, et al. Placental mitochondrial DNA and CYP1A1 gene methylation as molecular signatures for tobacco smoke exposure in pregnant women and the relevance for birth weight. J Transl Med. 2017;15(1):5.
Ezzeldin N, El-Lebedy D, Darwish A, El-Bastawisy A, Hassan M, Abd El-Aziz S, et al. Genetic polymorphisms of human cytochrome P450 CYP1A1 in an Egyptian population and tobacco-induced lung cancer. Genes Environ. 2017;39:7.
Fa S, Larsen TV, Bilde K, Daugaard TF, Ernst EH, Lykke-Hartmann K, et al. Changes in first trimester fetal CYP1A1 and AHRR DNA methylation and mRNA expression in response to exposure to maternal cigarette smoking. Environ Toxicol Pharmacol. 2018;57:19–27.
Vos S, Nawrot TS, Martens DS, Byun HM, Janssen BG. Mitochondrial DNA methylation in placental tissue: a proof of concept study by means of prenatal environmental stressors. Epigenetics. 2021;16(2):121–31.
Sharma H, Singh A, Sharma C, Jain SK, Singh N. Mutations in the mitochondrial DNA D-loop region are frequent in cervical cancer. Cancer Cell Int. 2005;5:34.
van Loon NM, Lindholm D, Zelcer N. The E3 ubiquitin ligase inducible degrader of the LDL receptor/myosin light chain interacting protein in health and disease. Curr Opin Lipidol. 2019;30(3):192–7.
Cardenas A, Lutz SM, Everson TM, Perron P, Bouchard L, Hivert MF. Mediation by placental DNA methylation of the association of prenatal maternal smoking and birth weight. Am J Epidemiol. 2019;188(11):1878–86.
Shorey-Kendrick LE, McEvoy CT, O’Sullivan SM, Milner K, Vuylsteke B, Tepper RS, et al. Impact of vitamin C supplementation on placental DNA methylation changes related to maternal smoking: association with gene expression and respiratory outcomes. Clin Epigenetics. 2021;13(1):177.
van Otterdijk SD, Binder AM, Michels KB. Locus-specific DNA methylation in the placenta is associated with levels of pro-inflammatory proteins in cord blood and they are both independently affected by maternal smoking during pregnancy. Epigenetics. 2017;12(10):875–85.
Rousseaux S, Seyve E, Chuffart F, Bourova-Flin E, Benmerad M, Charles MA, et al. Immediate and durable effects of maternal tobacco consumption alter placental DNA methylation in enhancer and imprinted gene-containing regions. BMC Med. 2020;18(1):306.
Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23(3):297–307.
Everson TM, Vives-Usano M, Seyve E, Cardenas A, Lacasana M, Craig JM, et al. Placental DNA methylation signatures of maternal smoking during pregnancy and potential impacts on fetal growth. Nat Commun. 2021;12(1):5095.
Song X, Wang Z, Zhang Z, Miao M, Liu J, Luan M, et al. Differential methylation of genes in the human placenta associated with bisphenol A exposure. Environ Res. 2021;200:111389.
Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018;145(11).
Ye Y, Tang Y, Xiong Y, Feng L, Li X. Bisphenol A exposure alters placentation and causes preeclampsia-like features in pregnant mice involved in reprogramming of DNA methylation of WNT2. FASEB J. 2019;33(2):2732–42.
Jedynak P, Tost J, Calafat AM, Bourova-Flin E, Busato F, Forhan A, et al. Pregnancy exposure to synthetic phenols and placental DNA methylation - an epigenome-wide association study in male infants from the EDEN cohort. Environ Pollut. 2021;290:118024.
Cao X, Hua X, Wang X, Chen L. Exposure of pregnant mice to triclosan impairs placental development and nutrient transport. Sci Rep. 2017;7:44803.
Wang X, Chen X, Feng X, Chang F, Chen M, Xia Y, et al. Triclosan causes spontaneous abortion accompanied by decline of estrogen sulfotransferase activity in humans and mice. Sci Rep. 2015;5:18252.
Kim S, Cho YH, Lee I, Kim W, Won S, Ku JL, et al. Prenatal exposure to persistent organic pollutants and methylation of LINE-1 and imprinted genes in placenta: a CHECK cohort study. Environ Int. 2018;119:398–406.
St-Pierre J, Hivert MF, Perron P, Poirier P, Guay SP, Brisson D, et al. IGF2 DNA methylation is a modulator of newborn’s fetal growth and development. Epigenetics. 2012;7(10):1125–32.
Constancia M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417(6892):945–8.
Zhao Y, Song Q, Ge W, Jin Y, Chen S, Zhao Y, et al. Associations between in utero exposure to polybrominated diphenyl ethers, pathophysiological state of fetal growth and placental DNA methylation changes. Environ Int. 2019;133(Pt B):105255.
Bro-Rasmussen F, Buus O, Trolle D. Ratio cortisone/cortisol in mother and infant at birth. Acta Endocrinol (Copenh). 1962;40:579–83.
Alikhani-Koopaei R, Fouladkou F, Frey FJ, Frey BM. Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J Clin Invest. 2004;114(8):1146–57.
Zhao Y, Gong X, Chen L, Li L, Liang Y, Chen S, et al. Site-specific methylation of placental HSD11B2 gene promoter is related to intrauterine growth restriction. Eur J Hum Genet. 2014;22(6):734–40.
Appleton AA, Jackson BP, Karagas M, Marsit CJ. Prenatal exposure to neurotoxic metals is associated with increased placental glucocorticoid receptor DNA methylation. Epigenetics. 2017;12(8):607–15.
Bromer C, Marsit CJ, Armstrong DA, Padbury JF, Lester B. Genetic and epigenetic variation of the glucocorticoid receptor (NR3C1) in placenta and infant neurobehavior. Dev Psychobiol. 2013;55(7):673–83.
Monk C, Feng T, Lee S, Krupska I, Champagne FA, Tycko B. Distress during pregnancy: epigenetic regulation of placenta glucocorticoid-related genes and fetal neurobehavior. Am J Psychiatry. 2016;173(7):705–13.
Seckl JR. Glucocorticoids, feto-placental 11 beta-hydroxysteroid dehydrogenase type 2, and the early life origins of adult disease. Steroids. 1997;62(1):89–94.
Gundacker C, Hengstschlager M. The role of the placenta in fetal exposure to heavy metals. Wien Med Wochenschr. 2012;162(9–10):201–6.
Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, Chen J, et al. Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of two U.S. birth cohorts. Environ Health Perspect. 2018;126(1):017010.
Tian FY, Everson TM, Lester B, Punshon T, Jackson BP, Hao K, et al. Selenium-associated DNA methylation modifications in placenta and neurobehavioral development of newborns: an epigenome-wide study of two U.S. birth cohorts. Environ Int. 2020;137:105508.
Kennedy E, Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, et al. Copper associates with differential methylation in placentae from two US birth cohorts. Epigenetics. 2020;15(3):215–30.
Le A, Shibata NM, French SW, Kim K, Kharbanda KK, Islam MS, et al. Characterization of timed changes in hepatic copper concentrations, methionine metabolism, gene expression, and global DNA methylation in the Jackson toxic milk mouse model of Wilson disease. Int J Mol Sci. 2014;15(5):8004–23.
Santos HP Jr, Bhattacharya A, Martin EM, Addo K, Psioda M, Smeester L, et al. Epigenome-wide DNA methylation in placentas from preterm infants: association with maternal socioeconomic status. Epigenetics. 2019;14(8):751–65.
Appleton AA, Armstrong DA, Lesseur C, Lee J, Padbury JF, Lester BM, et al. Patterning in placental 11-B hydroxysteroid dehydrogenase methylation according to prenatal socioeconomic adversity. PLoS ONE. 2013;8(9):e74691.
Clarkson-Townsend DA, Everson TM, Deyssenroth MA, Burt AA, Hermetz KE, Hao K, et al. Maternal circadian disruption is associated with variation in placental DNA methylation. PLoS ONE. 2019;14(4):e0215745.
Martin EM, Fry RC. A cross-study analysis of prenatal exposures to environmental contaminants and the epigenome: support for stress-responsive transcription factor occupancy as a mediator of gene-specific CpG methylation patterning. Environ Epigenet. 2016;2(1).
Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125(3):497–508.
Lyons AB, Moy L, Moy R, Tung R. Circadian rhythm and the skin: a review of the literature. J Clin Aesthet Dermatol. 2019;12(9):42–5.
Yuan V, Price EM, Del Gobbo G, Mostafavi S, Cox B, Binder AM, et al. Accurate ethnicity prediction from placental DNA methylation data. Epigenetics Chromatin. 2019;12(1):51.
Lee Y, Choufani S, Weksberg R, Wilson SL, Yuan V, Burt A, et al. Placental epigenetic clocks: estimating gestational age using placental DNA methylation levels. Aging (Albany NY). 2019;11(12):4238–53.
Funding
This work was supported by the National Institutes of Health (NIH-NIGMS T32GM008490, NIH-NIEHS R24ES028507, NIH-NIEHS R01ES025145, and NIH-NIEHS P30 ES019776).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Early Life Environmental Health.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mortillo, M., Marsit, C.J. Select Early-Life Environmental Exposures and DNA Methylation in the Placenta. Curr Envir Health Rpt 10, 22–34 (2023). https://doi.org/10.1007/s40572-022-00385-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40572-022-00385-1