Abstract
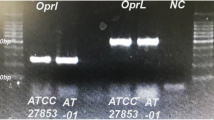
The soil and groundwater of Asanpara village (Bhagobangola I block) of Murshidabad district are contaminated with non-permissible limit of arsenic and other elements that co-exist with arsenic in various chemical compounds like arsenopyrite, ferrous arsenate, ferric arsenate, etc. Arsenic resistant bacteria (ARB) were isolated from arsenic contaminated soil of Asanpara and biochemically characterized. These bacteria were identified as Lysinibacillus sp. and Bacillus safensis by 16S rDNA sequencing and subsequent phylogenetic analysis. Isolated strains could grow in 76.98 mM and 88.53 mM of arsenite, and 560.88 mM and 721.13 mM of arsenate, respectively. ARB also show hypertolerance to other toxic metals like copper (Cu2+), cobalt (Co2+) and chromium (Cr3+). The level of arsenic and other heavy metal tolerance is unprecedented. Based on the inhibition of urease activity of isolated ARB by cadmium, the bacteria can be used to detect the presence of cadmium. These bacteria could biotransform arsenite into arsenate, which explains its uninhibited growth at very high arsenic concentration. The change in size of these bacteria depicted by scanning electron microscopy is a defence mechanism against arsenic stress Lysinibacillus sp. shows 32.33%, 31.29% and 31.20% bioremediation, whereas Bacillus safensis shows 37.54%, 35.26% and 35.24% bioremediation in the presence of 0.027 mM (2 ppm), 0.133 mM (10 ppm) and 0.667 mM (50 ppm) arsenic, respectively. Also, these bacteria could bioaccumulate or bioadsorb arsenic. The bioremediation potential of isolated ARB could be exploited for removal of arsenic from soil, groundwater and wastewater.








Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Availability of data and materials
Not applicable.
Code availability
Not applicable.
References
Abou-Shanaba RAI, Berkum PV, Angle JS (2007) Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere 68(2):360–367. https://doi.org/10.1016/j.chemosphere.2006.12.051
Altimira F, Yáñez C, Bravo G, González M, Rojas LA, Seeger M (2012) Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol 12:193. https://doi.org/10.1186/1471-2180-12-193
Banerjee S, Datta S, Chattyopadhyay D, Sarkar P (2011) Arsenic accumulating and transforming bacteria isolated from contaminated soil for potential use in bioremediation. J Environ Sci Health Part A 46:1736–1747. https://doi.org/10.1080/10934529.2011.623995
Bergey DH (1930) Bergey's Manual of Determinative Bacteriology: A Key for the Identification of Organisms of the Class Schizomycetes (3d ed.) 462p, Baltimore: Williams & Wilkins.
BIS (2012) Drinking water characteristics (IS:10500).
Brooks RR (1998) Plants That Hyperaccumulate Heavy Metals: Their Role in Phytoremediation, Microbiology, Archaeology, Mineral Exploration and Phytomining. 80p, Cambridge, University Press, USA. Publisher CAB International ISBN: 9780851991566 https://www.cabi.org/bookshop/book/9780851992365/
Brown AE, Benson HJ (2007) Benson’s Microbiological Applications. Laboratory Manual in General Microbiology, Short Version, 10 edn, The McGraw Hill Companies, New York, pp. 50
Camargo FAO, Okeke BC, Bento FM, Frankenberger WT (2005) Diversity of chromium-resistant bacteria isolated from soils contaminated with dichromate. Appl Soil Ecol 29(2):193–202. https://doi.org/10.1016/j.apsoil.2004.10.006
Cánovas D, Cases I, De Lorenzo V (2003) Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ Microbiol 5(12):1242–1256. https://doi.org/10.1111/j.1462-2920.2003.00463.x
Chakraborty M, Mukherjee A, Ahmed KM (2015) A Review of Groundwater Arsenic in the Bengal Basin, Bangladesh and India: from Source to Sink. Curr Pollution Rep 1:220–247. https://doi.org/10.1007/s40726-015-0022-0
Chandraprabha MN, Natarajan KA (2011) Mechanism of arsenic tolerance and bioremoval of arsenic by Acidithiobacilus ferrooxidans. J Biochem Tech 3(2):257–265. https://jbiochemtech.com/en/article/mechanism-of-arsenic-tolerance-and-bioremoval-of-arsenic-by-acidithiobacilus-ferrooxidans
Chowdhury R, Sen AK, Karak P, Chatterjee R, Giri AK, Chaudhuri K (2009) Isolation and characterization of an arsenic-resistant bacterium from a bore-well in West Bengal, India. Ann Microbiol 59:253–258. https://doi.org/10.1007/BF03178325
Coorevits A, Dinsdale AE, Heyrman J, Schumann P, Van Landschoot A, Logan NA, De Vos P (2012) Lysinibacillus macroides sp. nov., nom. rev. Int J Syst Evol Microbiol 62:1121–1127. https://doi.org/10.1099/ijs.0.027995-0
Dangleben NL, Skibola CF, Smith MT (2013) Arsenic Immunotoxicity: a Review. Environ Health 12(1):73. https://doi.org/10.1186/1476-069X-12-73
Dekker L, Osborne TH, Santini JM (2014) Isolation and identification of cobalt- and caesium-resistant bacteria from a nuclear fuel storage pond. FEMS Microbiol Lett 359(1):81–84. https://doi.org/10.1111/1574-6968.12562
Dey U, Chatterjee S, Mondal NK (2016) Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotechnol Rep. https://doi.org/10.1016/j.btre.2016.02.002
Dunivin TK, Miller J, Shade A (2018) Taxonomically-linked growth phenotypes during arsenic stress among arsenic resistant bacteria isolated from soils overlying the Centralia coal seam fire. PLoS ONE 13(1):e0191893. https://doi.org/10.1371/journal.pone.0191893
Dziubek J (2017) Arsenic removal from industrial wastewater. EKO-DOK 2017. E3S Web of Conferences 17, 00020. https://doi.org/10.1051/e3sconf/20171700020
Farooq SH, Chandrasekharam D, Dhanachandra W et al (2019) Relationship of arsenic accumulation with irrigation practices and crop type in agriculture soils of Bengal Delta, India. Appl Water Sci 9:119. https://doi.org/10.1007/s13201-019-0904-1
Fazi S, Amalfitano S, Casentine B et al (2015) Arsenic removal from naturally contaminated waters: a review of methods combining chemical and biological treatments. Rendiconti Lincei Scienze Fisiche e Naturali 27:51–58. https://doi.org/10.1007/s12210-015-0461-y
Fekih IB, Zhang C, Li YP, Zhao Y, Alwathnani HA, Saquib Q, Rensing C, Cervantes C (2018) Distribution of arsenic resistance genes in prokaryotes. Front Microbiol 9:2473. https://doi.org/10.3389/fmicb.2018.02473
Fitch WM, Margoliash E (1967) Construction of phylogenetic tree. Science 155(3760):279–284. https://doi.org/10.1126/science.155.3760.279
Gihring TM, Banfield JF (2001) Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol Lett 204(2):335–340. https://doi.org/10.1111/j.1574-6968.2001.tb10907.x
Goessler W, Kuehnett D (2002) Analytical methods for the determination of arsenic and arsenic compounds in the environment. Environmental chemistry of arsenic. In: Frankenberger Jr WTT (eds). Marcel Dekker, New York. Pp 27–50.
Guha Mazumder DN, Haque R, Ghosh N, De BK, Santra A, Chakraborty D, Smith AH (1998) Arsenic levels in drinking water and the prevalence of skin lesions in West Bengal. India Int J Epidemiol 27(5):871–877. https://doi.org/10.1093/ije/27.5.871
Gumpu MB, Sethuraman S, Krishnan UM, Rayappan JBB (2015) A review on detection of heavy metal ions in water—an electrochemical approach. Sens Actuators B Chem 213:515–533. https://doi.org/10.1016/j.snb.2015.02.122
Hao L, Mengzhu L, Wang N et al (2018) A critical review on arsenic removal from water using iron based adsorbents. Royal Soc Chem Adv. https://doi.org/10.1039/C8RA08512A
He M, Li X, Liu H, Miller SJ, Wang G, Rensing C (2011) Characterization and genomic analysis of a highly chromate resistant and reducing bacterial strain Lysinibacillus fusiformis ZC1. J Hazard Mater 185(2–3):682–688. https://doi.org/10.1016/j.jhazmat.2010.09.072
Hendryx M (2009) Mortality from heart, respiratory and kidney disease in coal mining areas of Appalachia. Int Arch Occup Env Health 82(2):243–249. https://doi.org/10.1007/s00420-008-0328-y
Igiri BE, Okoduwa SIR, Idoko GO, Akabuogu EP, Adeyi AO, Ejiogu IK (2018) Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. J Toxicol. https://doi.org/10.1155/2018/2568038
Islam K, Qian Qian W, Yu HJ, Chao W, Hua N (2017) Metabolism, toxicity and anticancer activities of Arsenic compounds. Oncotarget 8(14):23905–23926. https://doi.org/10.18632/oncotarget.14733
Jukes TH, Cantor CR (1969) Evolution of Protein Molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132. https://doi.org/10.1016/B978-1-4832-3211-9.50009-7
Kabir MS, Salam MA, Paul DN, Hossain MI, Rahman NM, Aziz A, Latif MA (2016) Spatial variation of arsenic in soil, irrigation water, and plant parts: a microlevel study. ScientificWorldJournal. https://doi.org/10.1155/2016/2186069
Kapaj S, Peterson H, Liber K, Bhattacharya P (2006) Human health effects from chronic Arsenic poisoning-a review. J Environ Sci Health Toxic Hazard Substan Environ Eng 41(10):2399–2428. https://doi.org/10.1080/10934520600873571
Krumova K, Nikolovska M, Groudeva V (2008) Isolation and Identification of Arsenic-Transforming Bacteria from Arsenic Contaminated Sites in Bulgaria. Biotechnol Biotechnol Equip 22(2):721–728. https://doi.org/10.1080/13102818.2008.10817541
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Lenoble V, Bouras O, Deluchat V, Serpaud B, Bollinger J (2002) Arsenic adsorption onto pillared clays and iron oxides. J Colloid Interface Sci 225:52–58. https://doi.org/10.1006/jcis.2002.8646
Maharjan M, Watanabe C, Ahmad SA, Umezaki M, Ohtsuka R (2007) Mutual interaction between nutritional status and chronic arsenic toxicity due to groundwater contamination in an area of Terai, lowland Nepal. J Epidemiol Community Health 61(5):389–394. https://doi.org/10.1136/jech.2005.045062
Marwa N, Singh N, Srivastava S, Saxena G, Pandey V, Singh N (2018) Characterizing the hypertolerance potential of two indigenous bacterial strains (Bacillus flexus and Acinetobacter junii) and their efficacy in arsenic bioremediation. J Appl Microbiol. https://doi.org/10.1111/jam.14179
McGowan W (2000) Water processing: residential, commercial, light-industrial, 3rd edn. Water Quality Association, Lisle.
Meharg AA, Hartley-Whitaker J (2002) Arsenic uptake and metabolism in arsenic resistant and non-resistant plant species. New Phytol 154:29–43. https://doi.org/10.1046/j.1469-8137.2002.00363.x
Mestrot A, Feldmann J, Krupp EM et al (2011) Field Fluxes and Speciation of Arsines Emanating From Soils. Environ Sci Technol 45:1798–1804. https://doi.org/10.1021/es103463d
Milton AH, Hasan Z, Shahidullah SM, Sharmin S, Jakariya MD, Rahman M, Dear K, Smith W (2004) Association between nutritional status and arsenicosis due to chronic arsenic exposure in Bangladesh. Int J Environ Health Res 4(2):99–108. https://doi.org/10.1080/0960312042000209516
Mondal NK, Roy P, Das B, Data JK (2011) Chronic arsenic toxicity and its relation with nutritional status: a case study in Purbasthali-II, Burdwan, west Bengal. Int J Environ Sci. https://doi.org/10.6088/ijes.00202020067
Mukherjee A, Fryar AE, Eastridge EM, Nally RS, Chakraborty M, Scanlon BR (2018) Controls on high and low groundwater arsenic on the opposite banks of the lower reaches of River Ganges, Bengal basin, India. Sci Total Environ 645:1371–1387. https://doi.org/10.1016/j.scitotenv.2018.06.376
Nicomel NR, Leus K, Folens K, Van Der Voort P, Laing GD (2016) Technologies for Arsenic Removal from Water: Current Status and Future Perspectives. Int J Environ Res Public Health 13(1):62. https://doi.org/10.3390/ijerph13010062
Oremland RS, Stolz JF (2003) The ecology of arsenic. Science 300:939–944. https://doi.org/10.1126/science.1081903
Páez-Espino D, Tamames J, de Lorenzo V, Cánovas D (2009) Microbial responses to environmental arsenic. Biometals 22(1):117–130. https://doi.org/10.1007/s10534-008-9195-y
Pais I, Benton Jons J (1997) The handbook of trace elements. St. Luice press Boca Rrton Florida. https://doi.org/10.2134/jeq1998.00472425002700040041x
Pakulska D, Czerczak S (2006) Hazardous effects of arsine: a short review. Int J Occup Med Environ Health 19(1):36–44. https://doi.org/10.2478/v10001-006-0003-z
Pandey N, Bhatt R (2015) Arsenic resistance and accumulation by two bacteria isolated from a natural arsenic contaminated site. J Basic Microbiol 55(11):1275–1286. https://doi.org/10.1002/jobm.201400723
Paul S, Majumdar S, Giri AK (2015) Genetic susceptibility to arsenic-induced skin lesions and health effects: a review. Genes and Environ 37:23. https://doi.org/10.1186/s41021-015-0023-7
Pelczar MJ, Bard RC, Burnett GW, Conn HJ, Demoss RD, Euans EE, Weiss FA, Jennison MW, Meckee AP, Riker AJ, Warren J, Weeks OB (1957) Manual of Microbiological Methods. Society of American Bacteriology, McGraw Hill Book Company, New York, pp. 315. https://doi.org/10.2307/1291949
Plewniak F, Crognale S, Rossetti S, Bertin PN (2018) A Genomic Outlook on Bioremediation: The Case of Arsenic Removal. Front Microbiol 9:820. https://doi.org/10.3389/fmicb.2018.00820
Prieto DM, González JC, Barral M (2018) Arsenic Mobility in As-Containing Soils from Geogenic Origin: Fractionation and Leachability. J Chem. https://doi.org/10.1155/2018/7328203
Rahman M, Vahter M, Sohel N, Yunus M, Wahed MA, Streatfield PK, Ekstrom EC, Persson LA (2006) Arsenic exposure and age and sex-specific risk for skin lesions: a population-based case-referent study in Bangladesh. Environ Health Perspect 114:1847–1852. https://doi.org/10.1289/ehp.9207
Rahman MM, Nag JC, Naidu R (2009) Chronic exposure of Arsenic via drinking water and its adverse health impacts on humans. Environ Geochem Health 31:189–200. https://doi.org/10.1007/s10653-008-9235-0
Rahman MM, Sengupta MK, Ahamed S, Lodh D, Das B, Hossain MA, Nayak B, Mukherjee A, Chakraborti D, Mukherjee SC, Pati S, Saha KC, Palit SK, Kaies I, Barua AK, Asad KA (2005) Murshidabad-One of the Nine Groundwater Arsenic-Affected Districts of West Bengal, India. Part I: Magnitude of Contamination and Population at Risk. Clin Toxicol 43:823–834. https://doi.org/10.1080/15563650500357461
Rahman A, Nahar N, Nawani NN, Jass J, Desale P, Kapadnis BP, Hossain K, Saha AK, Ghosh S, Olsson B, Mandal A (2014) Isolation and characterization of a Lysinibacillus strain B1-CDA showing potential for bioremediation of arsenics from contaminated water. J Environ Sci Health Part A Toxic/hazard Subst Environ Eng 49(12):1349–1360. https://doi.org/10.1080/10934529.2014.928247
Rahaman S, Sinha AC, Pati R, Mukhopadhyay D (2013) Arsenic contamination: a potential hazard to the affected areas of West Bengal. India Environ Geochem Health 35(1):119–132. https://doi.org/10.1007/s10653-012-9460-4
Raja EC, Omine K (2012a) Arsenic, boron and salt resistant Bacillus safensis MS11 isolated from Mongolia desert soil. Afr J Biotech 11(9):2267–2275. https://doi.org/10.5897/AJB11.3131
Raja EC, Omine K (2012) Characterization of boron resistant and accumulating bacteria Lysinibacillus sp. M1, Bacillus cereus M2, Bacillus cereus M3, Bacillus pumilus M4 isolated from former mining site, Hokkaido, Japan. J Environ Sci Health Part A Toxic/Hazard Substances Environ Eng 47(10):1341–1349. https://doi.org/10.1080/10934529.2012.672299
Ratnaike RN (2003) Acute and Chronic Arsenic Toxicity. Postgrad Med Jour 79(933):391–396. https://doi.org/10.1136/pmj.79.933.391
Roy P, Mondal NK, Das B, Das K (2013) Arsenic contamination in groundwater: a statistical modelling. Journal of Urban Environmental Engineering 7(1):24–29. https://doi.org/10.4090/juee.2013.v7n1.024029
Saha D, Sahu S (2016) A decade of investigations on groundwater arsenic contamination in Middle Ganga Plain. India Environ Geochem Health 38(2):315–337. https://doi.org/10.1007/s10653-015-9730-z
Saha JC, Dikshit AK, Bandyopadhyay M, Saha KC (1999) A review of Arsenic poisoning and its effects on human health. Crit Rev in Env Sci and Tech 29(3):281–313. https://doi.org/10.1080/10643389991259227
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Samal AC, Kar S, Maity JP, Santra SC (2013) Arsenicosis and its relationship with nutritional status in two arsenic affected areas of West Bengal, India. J Asian Earth Sci 77:303–310. https://doi.org/10.1016/j.jseaes.2013.07.009
Sankar MS, Vega MA, Defoe PP, Kibria MG, Ford S, Telfeyan K, Neal A, Mohajerin TJ, Hettiarachchi GM, Barua S, Hobson C, Johannesson K, Datta S (2014) Elevated arsenic and manganese in groundwaters of Murshidabad, West Bengal, India. Sci Total Environ 488–489:570–579. https://doi.org/10.1016/j.scitotenv.2014.02.077
Satomi M, La Duc MT, Venkateswaran K (2006) Bacillus safensis sp. nov., isolated from spacecraft and assembly-facility surfaces. Int J Syst Evol Microbiol 56:1735–1740. https://doi.org/10.1099/ijs.0.64189-0
Sen S, Saha T, Bhattacharya S, Nidhi, Mondal N, Ghosh W, Chakraborty R (2020) Draft Genome Sequences of Two Boron-Tolerant, Arsenic-Resistant, Gram-Positive Bacterial Strains, Lysinibacillus sp. OL1 and Enterococcus sp. OL5, Isolated from Boron-Fortified Cauliflower-Growing Field Soils of Northern West Bengal, India. Microbiology Resource Anouncements 9(2):e01438–19. https://doi.org/10.1128/MRA.01438-19
Shamim S (2018). Biosorption of Heavy Metals, Biosorption, Jan Derco and Branislav Vrana, IntechOpen. Available from: https://www.intechopen.com/chapters/58112https://doi.org/10.5772/intechopen.72099
Shen S, Li XF, Cullen WR, Weinfeld M, Le XC (2013) Arsenic Binding to Proteins. Chem Rev 113(10):7769–7792. https://doi.org/10.1021/cr300015c
Sher S, Rehman A (2019) Use of heavy metals resistant bacteria—a strategy for arsenic bioremediation. Appl Microbiol Biotechnol 103:6007–6021. https://doi.org/10.1007/s00253-019-09933-6
Shivaji S, Suresh K, Chaturvedi P, Dube S, Sengupta S (2005) Bacillus arsenicus sp. nov., an arsenic-resistant bacterium isolated from a siderite concretion in West Bengal. India Int J Syst Evol Microbiol 55:1123–1127. https://doi.org/10.1099/ijs.0.63476-0
Singh A (2006) Chemistry of arsenic in groundwater of Ganges–Brahmaputra river basin. Current Science, 91(5), 599–606. http://www.jstor.org/stable/24094363
Singh R, Singh S, Parihar P, Singh V, Prasad S (2015) Arsenic contamination, consequences and remediation techniques: A review. Ecotoxicol Environ Saf 112:247–270. https://doi.org/10.1016/j.ecoenv.2014.10.009
Smith AH, Hopenhayn-Rich C, Bates MN, Goeden HM, Hertz-Picciotto I, Duggan HM, Wood R, Kosnett MJ, Smith MT (1992) Cancer risks for Arsenic in drinking water. Environ Health Perspect 97:259–267. https://doi.org/10.1289/ehp.9297259
Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. WF Freeman & Co., San Francisco, p 573
SOES (2006) Groundwater arsenic contamination in West Bengal- India (20 years study), SOES.
Stolz JF, Basu P, Santini JM, Oremland RS (2006) Arsenic and selenium in microbial metabolism. Annu Rev Microbiol 60:107–130. https://doi.org/10.1146/annurev.micro.60.080805.142053
Stolz JF, Oremland RS (1999) Bacterial respiration of arsenic and selenium. FEMS Microbiol Rev 23:615–627. https://doi.org/10.1111/j.1574-6976.1999.tb00416.x
Syed S, Chinthala P (2015) Heavy Metal Detoxification by Different Bacillus Species Isolated from Solar Salterns. Scientifica (cairo) 2015:319760. https://doi.org/10.1155/2015/319760
Upadhyay N, Vishwakarma K, Singh J, Mishra M, Kumar V, Rani R, Mishra RK, Chauhan DK, Tripathi DK, Sharma S (2017) Tolerance and Reduction of Chromium(VI) by Bacillus sp. MNU16 Isolated from Contaminated Coal Mining Soil. Frontiers in Plant Science 8:778. https://doi.org/10.3389/fpls.2017.00778
Upadhyay MK, Yadav P, Shukla A and Srivastava S (2018) Utilizing the Potential of Microorganisms for Managing Arsenic Contamination: A Feasible and Sustainable Approach. Front. Environ. Sci. 6:24. https;//doi.org/https://doi.org/10.3389/fenvs.2018.00024
US EPA (2009) Drinking Water Requirements for States and Public Water System.
WHO (2011) Guidelines for drinking water quality. 4th edition. https://apps.who.int/iris/bitstream/handle/10665/44584/9789241548151_eng.pdf;jsessionid=F57D01D69337873BBBDMAB7A472A08DABF5?sequence=1
Xu S, Xu R, Nan Z, Chen P (2018) Bioadsorption of Arsenic from aqueous solution by the extremophilic bacterium Acidithiobacillus ferrooxidans DLC-5. Biocatal Biotransform 37(1):35–43. https://doi.org/10.1080/10242422.2018.1447566
Yoshida T, Yamauchi H, Sun GF (2004) Chronic health effects in people exposed to Arsenic via the drinking water: dose-response relationships in review. Toxicol Appl Pharmacol 198:243–252. https://doi.org/10.1016/j.taap.2003.10.022
Zapotoczna M, Riboldi GP, Moustafa AM, Dickson E, Narechania A, Morrissey JA, Planet PJ, Holden MTG, Waldron KJ, Geoghegan JA (2018) Mobile-Genetic-Element-Encoded Hypertolerance to Copper Protects Staphylococcus aureus from Killing by Host Phagocytes. mBio 9(5). https://doi.org/10.1128/mBio.00550-18
Zhu YG, Rosen BP (2009) Perspectives for genetic engineering for the photoremediation of arsenic contaminated environments: from imagination to reality? Curr Opin Biotechnol 20:220–224. https://doi.org/10.1016/j.copbio.2009.02.011
Acknowledgements
The authors acknowledge Department of Science and Technology and Biotechnology, Government of West Bengal, India, for funding the Research by its R&D Project Scheme—‘Gobeshonay Bangla’. The Biome Research Facility and Averin Biotech are acknowledged for their assistance in 16S rDNA sequencing and scanning electron microscopy analysis, respectively.
Funding
This work was supported by Department of Science and Technology and Biotechnology, Government of West Bengal, India [Grant No. STBT-11012(15)/26/2019-ST SEC].
Author information
Authors and Affiliations
Contributions
DM and AB conceived the idea and design of research. DM, RS and AB conducted the experiments. DM, RS, IS, SA and AB analysed the data. DM, IS, SA and AB were involved in manuscript preparation. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interests/competing interests of funding, employment, financial, non-financial or any other nature.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All the authors have given the consent for publication.
Additional information
Editorial responsibility: Jing Chen.
Rights and permissions
About this article
Cite this article
Mandal, D., Sonar, R., Saha, I. et al. Isolation and identification of arsenic resistant bacteria: a tool for bioremediation of arsenic toxicity. Int. J. Environ. Sci. Technol. 19, 9883–9900 (2022). https://doi.org/10.1007/s13762-021-03673-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-021-03673-9