Abstract
Purpose
Tumor cells thrive by adapting to the signals in their microenvironment. To adapt, cancer cells activate signaling and transcriptional programs and migrate to establish micro-niches, in response to signals from neighboring cells and non-cellular stromal factors. Understanding how the tumor microenvironment evolves during disease progression is crucial to deciphering the mechanisms underlying the functional behavior of cancer cells.
Methods
Multiplex immunohistochemistry, spatial analysis and histological dyes were used to identify and measure immune cell infiltration, cell signal activation and extracellular matrix deposition in low-grade, high-grade astrocytoma and glioblastoma.
Results
We show that lower grade astrocytoma tissue is largely devoid of infiltrating immune cells and extracellular matrix proteins, while high-grade astrocytoma exhibits abundant immune cell infiltration, activation, and extensive tissue remodeling. Spatial analysis shows that most T-cells are restricted to perivascular regions, but bone marrow-derived macrophages penetrate deep into neoplastic cell-rich regions. The tumor microenvironment is characterized by heterogeneous PI3K, MAPK and CREB signaling, with specific signaling profiles correlating with distinct pathological hallmarks, including angiogenesis, tumor cell density and regions where neoplastic cells border the extracellular matrix. Our results also show that tissue remodeling is important in regulating the architecture of the tumor microenvironment during tumor progression.
Conclusion
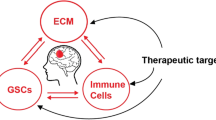
The tumor microenvironment in malignant astrocytoma, exhibits changes in cell composition, cell signaling activation and extracellular matrix deposition during disease development and that targeting the extracellular matrix, as well as cell signaling activation will be critical to designing personalized therapy.







Similar content being viewed by others
Data availability
Not applicable.
References
C. Wild, E. Weiderpass, B.W. Stewart, World cancer Report: cancer Research for cancer Prevention (IARC Press, 2020)
D.N. Louis, A. Perry, P. Wesseling, D.J. Brat, I.A. Cree, D. Figarella-Branger et al., The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23, 1231–1251 (2021)
D.N. Louis, A. Perry, G. Reifenberger, A. von Deimling, D. Figarella-Branger, W.K. Cavenee et al., The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131, 803–820 (2016)
R. Stupp, W.P. Mason, M.J. van den Bent, M. Weller, B. Fisher, M.J.B. Taphoorn et al., Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl. J. Med 352, 987–996 (2005)
H.S. Phillips, S. Kharbanda, R. Chen, W.F. Forrest, R.H. Soriano, T.D. Wu et al., Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006)
Cancer Genome Atlas Research Network, Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008)
C.W. Brennan, R.G.W. Verhaak, A. McKenna, B. Campos, H. Noushmehr, S.R. Salama et al., The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013)
S. Darmanis, S.A. Sloan, D. Croote, M. Mignardi, S. Chernikova, P. Samghababi et al., Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell. Rep 21, 1399–1410 (2017)
C.P. Couturier, S. Ayyadhury, P.U. Le, J. Nadaf, J. Monlong, G. Riva et al., Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun 11, 3406 (2020)
M. Castellan, A. Guarnieri, A. Fujimura, F. Zanconato, G. Battilana, T. Panciera et al., Single-cell analyses reveal YAP/TAZ as regulators of stemness and cell plasticity in glioblastoma. Nat. Cancer 2, 174–188 (2021)
L.M. Richards, O.K.N. Whitley, G. MacLeod, F.M.G. Cavalli, F.J. Coutinho, J.E. Jaramillo et al., Gradient of developmental and injury response transcriptional states defines functional vulnerabilities underpinning glioblastoma heterogeneity. Nat. Cancer 2, 157–173 (2021)
M. Rahman, J. Kresak, C. Yang, J. Huang, W. Hiser, P. Kubilis et al., Analysis of immunobiologic markers in primary and recurrent glioblastoma. J. Neurooncol 137, 249–257 (2018)
D. Yan, J. Kowal, L. Akkari, A.J. Schuhmacher, J.T. Huse, B.L. West et al., Inhibition of colony stimulating factor-1 receptor abrogates microenvironment-mediated therapeutic resistance in gliomas. Oncogene 36, 6049–6058 (2017)
B.M. Andersen, C. Faust Akl, M.A. Wheeler, E.A. Chiocca, D.A. Reardon, F.J. Quintana, Glial and myeloid heterogeneity in the brain tumour microenvironment. Nat. Rev. Cancer (2021). https://doi.org/10.1038/s41568-021-00397-3
M.G. García-Mendoza, D.R. Inman, S.M. Ponik, J.J. Jeffery, D.S. Sheerar, R.R. Van Doorn et al., Neutrophils drive accelerated tumor progression in the collagen-dense mammary tumor microenvironment. Breast Cancer Res 18, 49 (2016)
C.E. Barcus, P.Y. Hwang, V. Morikis, A. Brenot, P. Pence, M. Clarke et al. Tyrosine kinase-independent actions of DDR2 in tumor cells and cancer-associated fibroblasts influence tumor invasion, migration and metastasis. J. Cell. Sci. 134 (2021). https://doi.org/10.1242/jcs.258431
J.T. Rutka, C.A. Myatt, J.R. Giblin, R.L. Davis, M.L. Rosenblum, Distribution of extracellular matrix proteins in primary human brain tumours: an immunohistochemical analysis. Can. J. Neurol. Sci 14, 25–30 (1987)
H. Wang, Z. Liu, A. Li, J. Wang, J. Liu, B. Liu et al., COL4A1 as a novel oncogene associated with the clinical characteristics of malignancy predicts poor prognosis in glioma. Exp. Ther. Med 22, 1224 (2021)
L. MacCarthy-Morrogh, P. Martin. The hallmarks of cancer are also the hallmarks of wound healing. Sci. Signal. 13 (2020). https://doi.org/10.1126/scisignal.aay8690
P.M. Burkholder, Atlas of Human Glomerular Pathology: Correlative Light, Immunofluorescence, and Ultrastructural Histology (HarperCollins Publishers, 1974)
Hunter. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 9(03), 90–95 (2007)
J. Yuan, H.M. Levitin, V. Frattini, E.C. Bush, D.M. Boyett, J. Samanamud et al., Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med 10, 57 (2018)
A. Butler, P. Hoffman, P. Smibert, E. Papalexi, R. Satija, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018)
P.M. Daniel, G. Filiz, D.V. Brown, M. Christie, P.M. Waring, Y. Zhang et al., PI3K activation in neural stem cells drives tumorigenesis which can be ameliorated by targeting the cAMP response element binding protein. Neuro Oncol 20, 1344–1355 (2018)
D.F. Quail, J.A. Joyce, The microenvironmental landscape of brain tumors. Cancer Cell 31, 326–341 (2017)
J. Kowal, M. Kornete, J.A. Joyce, Re-education of macrophages as a therapeutic strategy in cancer. Immunotherapy 11, 677–689 (2019)
Cancer Genome Atlas Research Network, D.J. Brat, R.G.W. Verhaak, K.D. Aldape, W.K.A. Yung, S.R. Salama et al., Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl. J. Med 372, 2481–2498 (2015)
J.A. Miller, S.-L. Ding, S.M. Sunkin, K.A. Smith, L. Ng, A. Szafer et al., Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014)
A.M. Newman, C.L. Liu, M.R. Green, A.J. Gentles, W. Feng, Y. Xu et al., Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015)
A.J. Shaywitz, M.E. Greenberg, CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem 68, 821–861 (1999)
P.M. Daniel, G. Filiz, M.J. Tymms, R.G. Ramsay, A.H. Kaye, S.S. Stylli et al., Intratumor MAPK and PI3K signaling pathway heterogeneity in glioblastoma tissue correlates with CREB signaling and distinct target gene signatures. Exp. Mol. Pathol 105, 23–31 (2018)
S. Baumann, B. Kyewski, S.C. Bleckmann, E. Greiner, D. Rudolph, W. Schmid et al., CREB function is required for normal thymic cellularity and post-irradiation recovery. Eur. J. Immunol 34, 1961–1971 (2004)
K. Barton, N. Muthusamy, M. Chanyangam, C. Fischer, C. Clendenin, J.M. Leiden, Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB. Nature 379, 81–85 (1996)
B. Mastelic-Gavillet, B. Navarro Rodrigo, L. Décombaz, H. Wang, G. Ercolano, R. Ahmed et al., Adenosine mediates functional and metabolic suppression of peripheral and tumor-infiltrating CD8 + T cells. J. Immunother Cancer 7, 257 (2019)
F. Zhang, M. Rincon, R.A. Flavell, T.M. Aune, Defective th function induced by a dominant-negative cAMP response element binding protein mutation is reversed by Bcl-2. J. Immunol 165, 1762–1770 (2000)
K. Ohl, A. Schippers, K. Tenbrock, CD11c-Specific deletion reveals CREB as a critical regulator of DC function during the germinal center response. J. Immunol. Res 2018, 8947230 (2018)
B. Luan, Y.-S. Yoon, J. Le Lay, K.H. Kaestner, S. Hedrick, M. Montminy, CREB pathway links PGE2 signaling with macrophage polarization. Proc. Natl. Acad. Sci. U S A 112, 15642–15647 (2015)
L. Perria, U. Sacchi, [Incidence of age factor and tumoral stroma on the course of glioblastoma]. Sist Nerv 2, 176–186 (1950)
U. Novak, A.H. Kaye, Extracellular matrix and the brain: components and function. J. Clin. Neurosci 7, 280–290 (2000)
D.V. Brown, G. Filiz, P.M. Daniel, F. Hollande, S. Dworkin, S. Amiridis et al., Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS ONE 12, e0172791 (2017)
M. Ceccarelli, F.P. Barthel, T.M. Malta, T.S. Sabedot, S.R. Salama, B.A. Murray et al., Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016)
B. Weenink, K. Draaisma, H.Z. Ooi, J.M. Kros, P.A.E. Sillevis Smitt, R. Debets et al., Low-grade glioma harbors few CD8 T cells, which is accompanied by decreased expression of chemo-attractants, not immunogenic antigens. Sci. Rep 9, 14643 (2019)
F. Klemm, R.R. Maas, R.L. Bowman, M. Kornete, K. Soukup, S. Nassiri et al., Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181, 1643-1660.e17 (2020)
A.R. Pombo Antunes, I. Scheyltjens, F. Lodi, J. Messiaen, A. Antoranz, J. Duerinck et al., Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci 24, 595–610 (2021)
M.H. Robinson, J. Vasquez, A. Kaushal, T.J. MacDonald, J.E. Velázquez Vega, M. Schniederjan et al. Subtype and grade-dependent spatial heterogeneity of T-cell infiltration in pediatric glioma. J. Immunother Cancer. 8 (2020). https://doi.org/10.1136/jitc-2020-001066
K. Yan, Y. Lu, Z. Yan, Y. Wang, 9-Gene signature correlated with CD8 + T cell infiltration activated by IFN-γ: a biomarker of immune checkpoint therapy response in melanoma. Front. Immunol 12, 622563 (2021)
F. Basit, T. van Oorschot, J. van Buggenum, R.J.E. Derks, S. Kostidis, M. Giera et al., Metabolomic and lipidomic signatures associated with activation of human cDC1 (BDCA3+ /CD141+) dendritic cells. Immunology (2021). https://doi.org/10.1111/imm.13409
Y. Sun, A.J. Sedgwick, Y. Palarasah, S. Mangiola, A.D. Barrow, A transcriptional signature of PDGF-DD activated natural killer cells predicts more favorable prognosis in low-grade glioma. Front. Immunol 12, 668391 (2021)
Y.R. Na, J.W. Kwon, D.Y. Kim, H. Chung, J. Song, D. Jung et al., Protein kinase A catalytic subunit is a molecular switch that promotes the pro-tumoral function of macrophages. Cell. Rep 31, 107643 (2020)
X. Su, Y. Xu, G.C. Fox, J. Xiang, K.A. Kwakwa, J.L. Davis et al. Breast cancer-derived GM-CSF regulates arginase 1 in myeloid cells to promote an immunosuppressive microenvironment. J. Clin. Invest. 131 (2021). https://doi.org/10.1172/JCI145296
J.R.D. Pearson, T. Regad, Targeting cellular pathways in glioblastoma multiforme. Signal. Transduct. Target. Ther 2, 17040 (2017)
P.P. Provenzano, K.W. Eliceiri, J.M. Campbell, D.R. Inman, J.G. White, P.J. Keely, Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med (2006). https://doi.org/10.1186/1741-7015-4-38
A. Rambur, C. Lours-Calet, C. Beaudoin, J. Buñay, M. Vialat, V. Mirouse et al., Sequential Ras/MAPK and PI3K/AKT/mTOR pathways recruitment drives basal extrusion in the prostate-like gland of Drosophila. Nat. Commun 11, 2300 (2020)
Y.A. Yabo, S.P. Niclou, A. Golebiewska, Cancer cell heterogeneity and plasticity: a paradigm shift in glioblastoma. Neuro Oncol 24, 669–682 (2022)
Q. Xie, S. Mittal, M.E. Berens, Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol 16, 1575–1584 (2014)
H. Hatzikirou, D. Basanta, M. Simon, K. Schaller, A. Deutsch, “Go or grow”: the key to the emergence of invasion in tumour progression? Math. Med. Biol 29, 49–65 (2012)
Q.T. Ostrom, H. Gittleman, J. Xu, C. Kromer, Y. Wolinsky, C. Kruchko et al., CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol 18, v1–v75 (2016)
A. Garate-Carrillo, J. Gonzalez, G. Ceballos, I. Ramirez-Sanchez, F. Villarreal, Sex related differences in the pathogenesis of organ fibrosis. Transl Res 222, 41–55 (2020)
G. Escobar, B. Gentner, L. Naldini, R. Mazzieri, Engineered tumor-infiltrating macrophages as gene delivery vehicles for interferon-α activates immunity and inhibits breast cancer progression. Oncoimmunology 3, e28696 (2014)
L. Griffiths, K. Binley, S. Iqball, O. Kan, P. Maxwell, P. Ratcliffe et al., The macrophage - a novel system to deliver gene therapy to pathological hypoxia. Gene Ther 7, 255–262 (2000)
A.Y. Tsidulko, C. Bezier, G. de La Bourdonnaye, A.V. Suhovskih, T.M. Pankova, G.M. Kazanskaya et al., Conventional anti-glioblastoma chemotherapy affects proteoglycan composition of brain extracellular matrix in rat experimental model in vivo. Front. Pharmacol 9, 1104 (2018)
A. Zomer, D. Croci, J. Kowal, L. van Gurp, J.A. Joyce, Multimodal imaging of the dynamic brain tumor microenvironment during glioblastoma progression and in response to treatment. iScience 25, 104570 (2022)
A. Comba, S.M. Faisal, P.J. Dunn, A.E. Argento, T.C. Hollon, W.N. Al-Holou et al., Spatiotemporal analysis of glioma heterogeneity reveals COL1A1 as an actionable target to disrupt tumor progression. Nat. Commun 13, 3606 (2022)
Acknowledgements
We thank Metta Jana, Rejhan Idrizi and Ian Birchall for advice on tissue preparation, histology and staining. Marlene Hao and Lincon Stamp for reagents and scientific discussion.
Funding
This work was supported by the CASS Foundation Australia (grants 6236 and 7941) for T.M., Department of Surgery (RMH) and School of Biomedical Sciences translational research grant for T.M. and F.M.
Author information
Authors and Affiliations
Contributions
M.D. and S.S.W. performed most of the experimental work, data analysis and wrote parts of the manuscript. L.F. and L.C. performed immunohistochemical analysis experiments. Y.F. and S.M. performed the computational analysis experiments. P.N., P.D. and R.R. provided expertise on multiplex immunohistochemical analysis optimization experiments. F.M. provided expertise on tumor immunology and antibody panel selection. M.C. performed the histopathological analysis and IDH immunohistochemistry. S.S. provided expertise for glioma tissue selection and co-conceived the project. T.M. is the principal investigator who conceived the project, supervised research, and wrote and edited the manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Human ethics
Human ethics approval for tissue use was covered by project application 1853511 and was approved by the Medicine and Dentistry Human Ethics Sub-Committee, The University of Melbourne. For TMA specimens, all tissue was collected with patient and/or family consent.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1.53 MB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dinevska, M., Widodo, S.S., Furst, L. et al. Cell signaling activation and extracellular matrix remodeling underpin glioma tumor microenvironment heterogeneity and organization. Cell Oncol. 46, 589–602 (2023). https://doi.org/10.1007/s13402-022-00763-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-022-00763-9