Abstract
The main aim of this work was to induce the chromosome rearrangements between Aegilops ovata (UUMM) and hexaploid triticale (AABBRR) by expression of the gametocidal factor located on the chromosome 4M. The Aegilops ovata × Secale cereale (UUMMRR) amphiploids and triticale ‘Moreno’ were used to produce hybrids by reciprocal crosses. Chromosome dynamics was observed in subsequent generations of hybrids during mitotic metaphase of root meristems and first metaphase of meiosis of pollen mother cells. Chromosomes were identified by genomic in situ hybridisation (GISH) and fluorescence in situ hybridisation (FISH) using pTa71, pTa791, pSc119.2 and pAs1 DNA probes. It has been shown that the origin of the genetic background had an influence on Aegilops chromosome transmission. Moreover, it has been reported that the preferential transmission of chromosome 4M appeared during both androgenesis and gynogenesis. It is also hypothesised that the expression of the triticale Gc gene suppressor had an influence on the semi-fertility of hybrids but did not inhibit the chromosome rearrangements. This paper also describes the double haploid production, which enabled to obtain plants with two identical copies of triticale chromosomes with translocations of Aegilops chromatin segments.
Similar content being viewed by others
Introduction
Hexaploid triticale (×Triticosecale Wittm. ex A. Camus; 2n = 6x = 42, AABBRR) is a human-made cereal that was supposed to combine the yield potential of wheat (Triticum aestivum L.; 2n = 6x = 42, AABBDD) with the resistances and undemanding nature of rye (Secale cereale L.; 2n = 2x = 14, RR) in one plant. The breeding efforts aimed to develop a cereal adapted to dry locations, with higher protein content and a feeding quality similar to wheat. However, the synthetic character of this cereal resulted in narrow genetic variability. Moreover, the races of pathogens and pests managed to adapt to this new cereal and begun to break its resistance or tolerance abilities (Kwiatek et al. 2015a, b). To face this problem, there is a need to improve the genetic diversity of triticale by the transfer of desirable genes from wild relatives.
Distant crossing between alien species and wheat or triticale is often disturbed because of crossability barriers, such as different ploidy level of the parental components and the expression of the Ph1 gene (located on chromosome 5B), responsible for homologues chromosome pairing during meiosis (Riley and Chapman 1958; Griffiths et al. 2006; Lukaszewski and Kopecký 2010). There are several ways that induce the transfer of the chromosomes carrying the gene(s) of interests, including mutagenesis or the exploitation of the mechanisms that allow the homoeologous chromosomes to pair and recombine (absence of Ph1 gene or the presence of its recessive mutation ph1b), which are widely used to obtain the introgressive forms of wheat. The adaptation of the cereal recombinants into elite background is the crucial issue to deal with. Therefore, there is a need to find a natural process that will enable the chromosome segments of wild relatives to be transferred in a targeted manner into a wide range of cultivated species. In Aegilops species, a group of chromosomes called gametocidal (Gc) chromosomes are known to remain in wheat hybrid plants in a selfish way. The Gc chromosomes were first isolated during the production of alien cytoplasm substitution lines or alien chromosome addition lines in common wheat (Endo 1978). The expression of Gc factors brings on the sterility of gametes without Gc chromosomes, while the gametes with Gc chromosomes are fertile (Nasuda et al. 1998). Gc factors cause extensive chromosome breakage, which results in the induction of non-functional gametes and exclusive transmission of the Gc chromosome to the offspring (Nasuda et al. 1998). Sometimes, the Gc factor induces lower levels of chromosome breakage, which can be tolerated considering the polyploid nature of wheat and results in the formation of functional gametes (Nasuda et al. 1998). According to the literature, there are two phenotypes connected with the mechanism responsible for the preferential transmission of the Gc chromosomes (Endo 1990; Tsujimoto 2005). The first phenotype induces chromosome breakages, while the second prevents chromosome breakages. When both elements are present, the chromosome aberrations do not occur in gametes, because the gametocidal action is neutralised by the inhibitor. What is more, Friebe et al. (2003) reported the development of a knock-out wheat mutant carrying the Gc2 locus on Ae. sharonensis chromosome 4Ssh, which has lost the chromosome fragmentation function, but has retained the inhibitor element. The Gc system exploitation could be an effective way to induce chromosomal rearrangements and, what is better, it is conceived as safer and easier to handle than mutagens; however, the effectiveness of the Gc system in inducing gene mutations is not known (Endo 2007). Nonetheless, the molecular marker mapping enabled to localise Gc loci on a region proximal to a block of sub-telomeric heterochromatin on chromosome 4SshL of Ae. sharonensis (Knight et al. 2015). Gc chromosomes are derived from different genomes (C, S, Sl and Mg) and belong to three different homoeologous chromosome groups 2, 3 and 4 of the Aegilops genus (reviewed by Endo 2007).
Aegilops ovata Roth. (syn. Ae. geniculata; van Slageren 1994) is a tetraploid (2n = 4x = 28, UgUgMgMg). The Ug genome originated from the U genome of the diploid species Ae. umbellulata Zhuk. (2n = 2x = 14, UU) and the Mg genome was derived from the M genome of Ae. comosa Sm. in Sibth. & Sm. (2n = 2x = 14, MM) (Kihara 1954). Aegilops ovata has a wide distribution and is native to the Mediterranean, Middle East and southern parts of Russia and Ukraine. The Gc factor was localised on the 4Mg chromosome (Kynast et al. 2000). Aegilops ovata is a valuable source of resistance genes for biotic and abiotic stresses (Schneider et al. 2008), which can be used for improving cultivated cereals, such as wheat (Friebe et al. 2000), rye (Apolinarska et al. 2010; Kwiatek et al. 2012) and triticale (Kwiatek et al. 2015a, b).
The main aim of this work was to induce the chromosome translocations between Ae. ovata (UUMM) and triticale (AABBRR) genomes by the gametocidal system. In this paper, we present an evaluation of the transmission rate of Aegilops chromatin in the triticale genomic background. We have used Ae. ovata × S. cereale amphiploids as parental forms to cross with triticale. The double haploid lines were obtained from fertile or semi-fertile F2 hybrids in order to stabilise the Aegilops chromatin in the triticale background. Genomic/fluorescence in situ hybridisation (GISH/FISH) techniques were applied to identify the genomes and particular chromosomes.
Materials and methods
Plant material
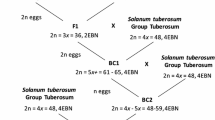
Glasshouse experiments were carried out at the Institute of Plant Genetics, Polish Academy of Sciences in Poznań, Poland. Seeds of Ae. ovata (To36; 2n = 4x = 28; UUMM) were received from the collection of Professor Moshe Feldman (Department of Plant Science, The Weizmann Institute of Science, Rehovot, Israel). The Ae. ovata × S. cereale amphiploids (UUMMRR, 2n = 6x = 42) were obtained by Wojciechowska and Pudelska (2002). The F1 (Ae. ovata × S. cereale) × triticale hybrids were obtained by reciprocal crossing with triticale cv. ‘Moreno’ received from DANKO Breeding Company (DANKO Hodowla Roślin Sp. z o.o., Choryń, Poland). The following steps and results of offspring production are described in Fig. 1 and Table 1.
Evaluation of pollen vitality, crossing and crossing efficiency of hybrids
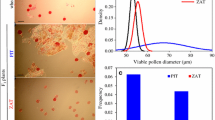
The pollen grains were stained with 2 % acetocarmine in glycerine (vol. 1:1) for the presence of cytoplasm. The evaluation of pollen vitality was made using an Olympus BX60 microscope.
Maternal components were emasculated to avoid self-fertilisation in order to cross with the pollen of triticale cv. ‘Moreno’. The emasculated flowers were counted and pollinated with freshly collected pollen of triticale within a period of 3 months (April–June). The percentage ratio of the total amount of seeds from each plant with the total amount of pollinated flowers of each plant was calculated. The mean of the crossing efficiency of each generation of hybrids was established.
Double haploid production
The tillers from donor plants were cut at the microspore stage and cold treated at 4 °C for 6–9 days in mineral salt medium N6 (Chu et al. 1975) +2.0 mg/l 2,4-D in the dark. The anthers were cultured on the C17 medium (Wang and Chen 1983) +2 mg/l 2,4-D + 0.5 mg/l KIN + 90 g/l of maltose in darkness at 28 °C. Androgenic structures developed from the microspores were placed in the regeneration medium 190–2 (Zhuang and Xu 1983). Regeneration was induced at 22 °C, in continuous light for 12 h per day. The ploidy level was evaluated on the basis of gDNA content in leaves tissue using flow cytometry. Chromosome doubling was obtained using 0.1 % colchicine solution supplemented with 4 % DMSO and 25 mg/l GA3. Afterwards, the plants were placed in the vernalisation chamber for six weeks and then located in the glasshouse until harvest.
Chromosome preparation
Germination, metaphase accumulation and fixation procedures were carried out according to Kwiatek et al. (2015a). Maceration was made in 0.2 % (v/v) Onozuka R-10 and Calbiochem cytohelicases (1:1 ratio) and 20 % pectinase (Sigma) in 10 mM citrate buffer (pH 4.6) at 37 °C for 30 min and stopped by washing two times for 10 min each in citrate buffer and in H2O. Chromosome preparations were made according to the protocol reported by Heckmann et al. (2014), with modifications made by Hesse (IPK Gatersleben, Germany, personal comm.). Root tips were placed on a slide in a drop of ice-cold 60 % acetic acid and dispersed with a metal needle. Ice-cold acetic acid (60 %) was added to the cell suspension, mixed and kept for 2 min at room temperature. Next, ice-cold 60 % acetic acid was added and the slide was placed on a heating table (Medax) for 2 min at 48 °C. The slide was removed from the hot plate and 200 μl ice-cold ethanol–acetic acid (3:1, v/v) was added to briefly wash the slides. The slide was placed in 60 % acetic acid for 10 min, followed by briefly washing in 96 % ethanol and then air dried. Hybrids were grown in the nursery and their meiotic behaviour was analysed in pollen mother cells (PMCs) at metaphase I (MI) of meiosis. Anthers of the hybrids containing PMCs at MI were fixed in 1:3 (v/v) acetic acid/ethanol and stored at −20 °C for a maximum of 2 months. MI of meiosis preparations were made according to Zwierzykowski et al. (2008). The anthers were squashed in 45 % acetic acid and the slides were stored at 4 °C until in situ hybridisation.
Probe labelling
Total genomic DNA was extracted from fresh leaves of Ae. umbellulata (UU), Ae. comosa (MM) and triticale ‘Moreno’ (AABBRR) using GeneMATRIX Plant & Fungi DNA Purification Kit (EURx Ltd.). Genomic DNA from Ae. umbellulata and Ae. comosa were labelled by nick translation (using the Nick Translation Kit, Roche, Mannheim, Germany) with digoxigenin-11-dUTP (Roche) or tetramethyl-5-dUTP-rhodamine (Roche), respectively. Blocking DNA from triticale was sheared by boiling for 30–45 min and used at a ratio of 1:50 (probe:block). The 5S rDNA probe was amplified from the wheat clone pTa794 (Gerlach and Dyer 1980) by polymerase chain reaction (PCR) with tetramethyl-rhodamine-5-dUTP (Roche) using universal M13 ‘forward’ (5′-CAG GGT TTT CCC AGT CAC GA-3′) and ‘reverse’ (5′-CGG ATA ACA ATT TCA CAC AGG A-3′) sequencing primers. The PCR reactions were performed as follows: 94 °C for 1 min, 39 cycles of 94 °C for 40 s, 55 °C for 40 s and 72 °C for 90 s, and 72 °C for 5 min. The 25S rDNA probe was made by nick translation of a 2.3-kb ClaI sub-clone of the 25-5.8-18S rDNA coding region of Arabidopsis thaliana (Unfried and Gruendler 1990) with digoxigenin-11-dUTP (Roche). It was used for the detection of 25-5.8-18S rDNA loci. The pSc119.2 repetitive DNA sequence, supplied by Dr. Kubalaková (Laboratory of Molecular Cytogenetics and Cytometry, Institute of Experimental Botany, Olomouc, Czech Republic), was amplified and labelled by PCR with digoxigenin-11-dUTP (Roche) using universal M13 primers (Vrána et al. 2000). The pAs1 probe was amplified by PCR from the genomic DNA of Ae. tauschii and labelled with digoxigenin-11-dUTP (Roche), according to the protocol reported by Nagaki et al. (1995).
In situ hybridisation
GISH was carried out according to Kwiatek et al. (2012), with modifications. Multi-colour GISH was carried out using the U-genome probe (from Ae. umbellulata), M-genome probe (from Ae. ovata) and unlabelled triticale genomic DNA, which was used as a specific blocker. The GISH mixture (40 μl per slide), containing 50 % formamide, 2 × SSC, 10 % dextran sulphate, 90 ng each of the genome probes and 4.5 μg blocking DNA, was denatured at 75 °C for 10 min and stored on ice for 10 min. Chromosomal DNA was denatured in the presence of the hybridisation mixture at 75 °C for 5 min and allowed to hybridise overnight at 37 °C. For detection of the hybridisation signals, anti-digoxigenin IgG conjugated with FITC (Roche) was used. After documentation of the FISH sites, the slides were washed according to Heslop-Harrison (2000) (2 × 45 min in 4 × SSC Tween, 2 × 5 min in 2 × SSC, at room temperature). To recognise the particular chromosomes, we used four probes for in situ hybridisation on the same chromosome preparations in two subsequent FISH runs. The first FISH was carried out according to Książczyk et al. (2011), with minor modifications of Kwiatek et al. (2013), using 25S (used for the detection of 25-5.8-18S rDNA loci) and 5S rDNA (pTa794). The hybridisation mixture (40 μl per slide) contained 90 ng of each probe in the presence of salmon sperm DNA, 50 % formamide, 2 × SSC and 10 % dextran sulphate, and was denatured at 75 °C for 10 min and stored on ice for 10 min. The second FISH, with pSc119.2 and pAs1 (labelled with digoxigenin-11-dUTP and tetramethyl-rhodamine-5- dUTP, respectively), was performed with the same conditions after reprobing. The chromosomal DNA denaturation, hybridisation and immunodetection conditions were the same as those mentioned above. Digoxigenin detection was carried out using anti-digoxigenin-fluorescein antibody (Roche). FISH was carried out to study the mitotic chromosomes of root meristems. On the other hand, GISH was used to examine both the mitotic chromosomes of root meristems and meiotic chromosomes of PMCs. Mitotic and meiotic (MI) cells were examined with an Olympus XM10 CCD camera attached to an Olympus BX 61 automatic epifluorescence microscope. Image processing was carried out using Olympus Cell-F (version 3.1; Olympus Soft Imaging Solutions GmbH, Münster, Germany) imaging software and PaintShop Pro X5 software (version 15.0.0.183; Corel Corporation, Ottawa, Canada). The identification of particular chromosomes was made by comparing the signal pattern of 5S rDNA, 25S rDNA, pSc119.2 and pAs1 probes according to a previous study (Kwiatek et al. 2013) and similar cytogenetic analysis (Cuadrado and Jouve 1994; Schneider et al. 2003, 2005; Wiśniewska et al. 2013).
Results
Evaluation of pollen viability and crossing efficiency
The mean percentage of pollen viability of Ae. ovata × S. cereale amphiploids was 64.4 %, while the pollen viability of triticale var. ‘Moreno’ ranged from 86 to 94 %. Reciprocal hybridisation was done to obtain hybrid forms between triticale ‘Moreno’ (AABBRR) and Ae. ovata × S. cereale (UUMMRR) amphiploids. Triticale pollen was used to pollinate 168 flowers of Ae. ovata × S. cereale plants. Twenty-eight obtained seeds were germinated, self-pollinated and back-crossed with triticale. The mean pollen viability of (Ae. ovata × S. cereale) × triticale F1 hybrids was 4.82 %. 13 F2 and 0 BC1F1 seeds were harvested after self-pollinating or back-crossing of 212 and 468 flowers, respectively. The pollen viability of F2 plants was 32.42 %. Self-pollination of 334 flowers enabled to obtain 93 seeds of the F3 generation. Simultaneously, 354 flowers of triticale ‘Moreno’ were pollinated by the pollen of Ae. ovata × S. cereale forms. Four F1 seeds were germinated and evaluated using FISH/GISH analysis. The F1 plants produced pollen grains with 9.09 % viability. Self-pollination of 248 flowers resulted in 16 seeds of F2 progeny. Back-crossing of 334 flowers of F1 hybrids with the triticale ‘Moreno’ pollen gave 71 seeds of the BC1F1 hybrid generation. The mean pollen viability was 24.20 % for F2 hybrids and 50.93 % for BC1F1 plants. Afterwards, the F2 hybrids (381 flowers) were self-pollinated and BC1F1 plants (1518 flowers) were pollinated with triticale pollen. 56 F3 and 1058 BC2F1 seeds were harvested, respectively.
Anther cultures in Aegilops triticale hybrids
Overall, 2896 anthers from F3 ‘Moreno’ × (Ae. ovata × S. cereale) plants and 3204 anthers from BC2F1 hybrids were used to produce 3461 (119.5 per 100 anthers) and 4444 (138.7 per 100 anthers) androgenic structures, from which 18 (0.62) and 40 (1.3 per 100 anthers) haploid plants were obtained, respectively. Thereafter, 6 and 17 double haploid lines (respectively) of introgressive triticale were generated (Fig. 1) using the colchicine solution to double the ploidy level.
Transmission of Aegilops ovata chromatin in triticale hybrids
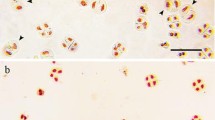
The chromosome constitution of both F1 (triticale as a male or female parent) combinations was the same and consist of: 14 chromosomes of Ae. ovata (seven chromosomes of U-genome and seven chromosomes of M-genome) and 28 chromosomes of triticale (14 chromosomes of R-genome and 14 chromosomes of A- and B-genomes; Table 1; Fig. 2a). The total number of chromosomes was 42. In 13 F2 plants of the (Ae. ovata × S. cereale) × triticale combination, the number of chromosomes ranged from 38 to 41. The range of both U- and M-genome chromosomes was 2–4. In each of the 13 plants, a pair of 4M chromosomes was identified (Table 2; Fig. 2c, d). Chromosomes 1U, 5U, 7U, 1M, 6M and 7M were eliminated.
a Genomic in situ hybridisation (GISH) experiment showing seven U-genome (red) and seven M-genome (green) chromosomes of Aegilops ovata and 28 triticale chromosomes (grey and blue) of F1 plants during metaphase I (MI) of mitosis. b GISH experiment showing mitotic chromosomes of U-genome (red), M-genome (green) and triticale genomes (grey and blue) in F2 triticale × (Ae. ovata × Secale cereale) plants. c Fluorescence in situ hybridisation (FISH) experiment presenting the location of pSc119.2 (green) and pAs1 (red) probes in mitotic chromosomes of F2 (Ae. ovata × S. cereale) × triticale plants. d GISH experiment showing chromosomes of U-genome (red), M-genome (green) and triticale genomes (grey and blue) during mitosis of the same F2 (Ae. ovata × S. cereale) × triticale plants. Mitotic chromosomes of BC1F1 triticale × (Ae. ovata × S. cereale) plants analysed by: e FISH with 5S and 35S rDNA probes, f FISH with pSc119.2 (green) and pAs1 (red) probes and g GISH with U-genome (red), M-genome (green) and triticale (grey and blue) genomic probes. h F1 plant during MI of meiosis carrying M-genome bivalent and UMU-genomes trivalent (arrows) and i F2 (Ae. ovata × S. cereale) × triticale plant carrying 4M chromosome bivalent (arrow). Scale bar: 10 μm
On the other hand, the chromosome constitution of F2 triticale × (Aegilops ovata × S. cereale) hybrids was different. The total number of chromosomes ranged between 41 and 42. The lack of Aegilops chromosomes was revealed in three plants. The range of Aegilops chromosomes in the remaining 15 plants was 0–3 and 0–5 for U- and M-genomes, respectively (Fig. 2b). The most common were 4M and 2M chromosomes. In three plants, a pair of 4M chromosomes was identified. Aegilops chromosomes from the 1st, 6th and 7th homoeologous groups were not identified (Table 2).
Complete Aegilops chromosomes were absent in each of the 71 BC1F1 plants of the triticale × (Ae. ovata × S. cereale) combination; however, each of the A- and B-genome chromosomes of triticale carried small segments or distant signals on M-genome chromatin, except for the 5A chromosome pair. Clear M-genome probe signals were also detected on chromosomes of the 2R pair (Fig. 2e, f, g). The total number of chromosomes in BC1F1 plants was 40 (in 23 plants), 41 (12 plants) and 42 (36 plants). In the nullisomic or monosomic plants of triticale hybrids, the 2A, 2B or 3B chromosome pairs or singe chromosomes were missing.
The chromatin of Ae. ovata was completely eliminated in 50 F3 plants of the (Ae. ovata × S. cereale) × triticale combination. Those plants carried different numbers of triticale chromosomes, ranging between 34 and 42. The 2B, 3B, 4B and 6B chromosomes or chromosome pairs were lacking in these forms. Complete Ae. ovata chromosomes were also absent in 56 F3 and 50 BC2F1 plants of the triticale × (Aegilops ovata × S. cereale) combination. However, the chromosomes of A- and B-genomes carried M-genome chromatin translocations, which were similar to those observed in BC1F1 plants.
Finally, the 12 and 34 DH plants obtained from F3 and BC2F1 hybrids of the triticale × (Aegilops ovata × S. cereale) combination carried 42 chromosomes. The most abundant signals of the M-genome probe appeared on 1A, 2A, 1B, 3B, 4B, 6B and in the short arm of 2B. The M-genome chromosome segments were located in pericentromeric regions on the 2A, 4B and 6B chromosomes of triticale (Fig. 3). Clear but single signals were observed on the 3A, 4A, 6A, 7A, 5B, 7B and 2R chromosomes as well (Fig. 4). Moreover, M-genome landmarks were dispersed on the 1A, 1B and 3B chromosomes. Furthermore, M-genome signals were also observed in telomeric regions of the 7AS, 1BS, 1BL, 2AL, 3BL, 4BL and 6BL chromosome arms.
Aegilops ovata chromosome behaviour at metaphase I of meiosis of F1, F2 and BC1F1 hybrids
The meiotic analyses were done in two initial generations of hybrids, where Ae. ovata chromosomes were established. The chromosome configurations of 32 F1 plants during MI of meiosis were in parallel with the theoretical assumptions. Seven univalents of the U-genome as well as seven univalents of the M-genome were observed. The number of A- and B-genome chromosomes was haploid as well. Furthermore, the R-genome chromosomes paired and created rod or ring bivalents. However, in two other F1 plants from the (Ae. ovata × S. cereale) × triticale combination, one ring bivalent of the M-genome was detected. Moreover, those hybrids also carried a trivalent that consisted of two U-genome chromosomes linked with one M-genome chromosome (Table 3; Fig. 2h). In the F2 generation of the (Ae. ovata × S. cereale) × triticale combination, the number of Ae. ovata univalents ranged between 0 and 8. One ring bivalent of the M-genome was detected (Fig. 2i). Moreover, the diverse types of intergenomic bivalents, trivalents and tetravalents were observed. The frequencies are shown in Table 3. On the other hand, the Ae. ovata univalents were more abundant in the F2 generation of the triticale × (Ae. ovata × S. cereale) combination, while the frequencies of bivalents and multivalents were lower (Table 3). What is more, the M-genome bivalents were not observed.
Discussion
Considering the narrow genetic diversity of triticale, it is necessary to utilise the wild Triticeae relatives to enrich the genetic pool of this species. The most common method used to transfer genes of agronomic value from wild relatives into cultivated cereals are interspecific hybridisations. Initially, one of the main breeding strategies aiming at triticale improvement was to introduce D-genome chromosomes into 6x triticale. A number of efforts in the production of triticale substitution lines were conducted. The simplest way to obtain the D(A) or/and D(B) substitution lines is an application of octoploid (AABBDDRR) × tetraploid (AARR or BBRR) triticale crosses (Krowlow 1970; Lukaszewski et al. 1984; Apolinarska 1993). Moreover, as well as in wheat breeding, several attempts were made to cross triticale with Aegilops species (Gruszecka et al. 1996; Gradzielewska et al. 2012; Kwiatek et al. 2015a, b), which are related to Triticum species and represent a large source of useful traits for crop improvement (Schneider et al. 2008). The alien species Ae. speltoides, Ae. cylindrica, Ae. sharonensis, Ae. longissima and Ae. ovata are reported to posses gametocidal genes, which give rise to chromosomal rearrangements (Endo 2007).
Up to now, the process of preferential transmission of chromosomes carrying gametocidal (Gc) factors that originated from Aegilops species were reported only considering the wheat genetic background (Finch et al. 1984; Friebe et al. 1999; Kynast et al. 2000; Endo 2007; Ishihara et al. 2014). Here, we report novel data about the behaviour of Ae. ovata chromosomes in the triticale genetic background. It is worth mentioning that Aegilops chromosomes were transferred to triticale from a stable and fertile (64.4 %) Ae. ovata × S. cereale amphiploid form, where Ae. ovata was a female parent (Wojciechowska and Pudelska 2002). Therefore, the cytoplasm of the amphiploid originated from Aegilops species. Hence, the reciprocal crosses between Aegilops–Secale forms and triticale were made to evaluate the impact of the cytoplasm on the transmission of the Ae. ovata chromatin to hybrid progeny.
All of the F1 plants obtained from both combinations carried the expected haploid number of Aegilops chromosomes. These hybrids underwent meiosis, showing, in most cases, 14 univalents of Ae. ovata, 14 univalents from A- and B-genomes, and 14 bivalents from the R-genome. However, pairing of M-genome chromosomes was observed in the pollen mother cells of two plants. Additionally, another M-genome chromosome was involved in trivalent formation with two U-genome chromosomes. This phenomena seems to be cryptic, considering that each of the Aegilops chromosomes appeared in one copy during the MI of F1 hybrids. It could be possible that homoeologous pairing took place to balance the lack of homologues. The F2 and BC1F1 hybrids were obtained, regardless of chromosome configuration during the MI of meiosis, which caused low pollen fertility as well as decreased crossing ability of F1 hybrids.
All of the F2 plants with Aegilops cytoplasm carried a pair of 4M chromosomes, which suggested that the preferential transmission of that particular chromosome appeared during both androgenesis and gynogenesis. What is more, those chromosomes paired during the MI of meiosis of each F2 plant PMCs (Table 3; Fig. 2i). In contrast, the process of preferential transmission was weaker considering the F2 progeny carrying triticale cytoplasm. In this case, only three plants carried a pair of 4M chromosomes. The rest of the F2 plants possess one copy of the 4M chromosome. Furthermore, three other plants did not carry any of the Aegilops chromosomes (Table 2). Both combinations gave semi-fertile pollen grains, but the pollen viability of F2 progeny carrying Aegilops cytoplasm (32.42 %) was higher than in the F2 hybrids of triticale × (Ae. ovata × S. cereale).
On the other hand, back-crossing with the triticale pollen resulted in a lack of BC1F1 hybrids with Aegilops cytoplasm. In contrast, the same crossing performed on F1 hybrids with triticale cytoplasm effected many translocations of Aegilops chromatin in triticale chromosomes. It could be supposed that the origin of the cytoplasm has an influence on Aegilops chromosome transmission. This result also enable to assume the existence of a cytoplasmic factor that increases the effect of 4M chromosome transmission on one hand and causes abortion of a gamete lacking the 4M chromosome on the other. On the contrary, the existence of Gc suppressor, which decreases the effect of preferential transmission and makes the gametes lacking the 4M chromosome fertile or semi-fertile, could be considered as a counter-hypothesis. This point of view is in parallel to the observations made by Tsujimoto and Tsunewaki (1985) considering the different level of PMCs viability of monosomic wheat: ‘Chinese Spring’, ‘John Fife’ and ‘Norin26’ carrying additional an Gc chromosome 3C of Ae. triuncialis. The first two forms showed both male and female semi-sterility (Endo 1978). However, semi-sterility did not appear in the cultivar ‘Norin26’. Moreover, chromosome 3C was preferentially transmitted to the next generation from both paternal and maternal sides in ‘John Fife’, but only from the female side in ‘Chinese Spring’. In the ‘Norin26’ cultivar, the chromosome 3C was transmitted normally as a usual alien monosome without preferential transmission (Endo 1978). Tsujimoto and Tsunewaki (1985) crossed the ‘Chinese Spring’ addition line carrying two 3C chromosomes with the F1 progeny of ‘Chinese Spring’ and ‘Norin26’ and obtained the monosomic addition lines, fertile and semi-sterile plants segregated 1:1. On this basis, they suspected that a dominant suppressor gene (Igc1) controls the suppression of Gc gene expression on chromosome 3C. In our study, the genetic background of cultivar ‘Moreno’ provided the semi-sterility of PMCs but did not prevent the fragmentation of Aegilops chromosomes in F3, BC1F1 and BC2F1 plants. On this basis, it can be assumed that the suppressing factor showed incomplete expression.
Analogous conclusions were formed by Tsujimoto and Tsunewaki (1985), who reported that, in the ‘John Fife’ genetic background, both male and female gametes without chromosome 3C were abortive, while in the ‘Chinese Spring’ genetic background, pollen without the Gc chromosome functioned and transmitted to the progeny, indicating the expression of an incomplete suppressor in the ‘Chinese Spring’ cytoplasm. The M-genome translocations on chromosomes of F3 triticale were located in the pericentromeric region and located on chromosomes 1A, 4B and 6B. The rest of the signals, which were observed on A-, B- and R-genome chromosomes, probably show the distribution of repetitive sequences that are specific for Triticeae species. The existence of classes of widespread repetitive sequences have been reported in the tribe Triticeae (Vershinin et al. 1994). The occurrence of those signals, detected in distant hybrids, indicates the high level of resemblance between the repetitive sequences in selected species. More detailed analysis should be made to establish the precise locations of M-genome chromatin in triticale chromosomes.
The patterns of M-genome signals were similar in double haploid lines obtained from F3 and BC2F1. The effectiveness of in vitro cultures, treated as the number of plants compared to the number of anthers, is a crucial factor for producing prebreeding germplasm. The number of green haploid plants was 18 for F3 (0.62 per 100 anthers) and 40 for BC2F1 (1.3 per 100 anthers), and was low when compared to similar experiments carried out on triticale, which resulted from 0.4 to even 7.9 green plants per 100 anthers (Ślusarkiewicz-Jarzina and Ponitka 1997, 2003; Ponitka et al. 1999). Double haploids are being produced from triticale in support of breeding programmes. Haploid lines and haploidy on intergeneric hybrids have been successively applied to bridge the D- and R-genomes in the 6x triticale and for creating alien addition lines of F1 hybrids (Wang and Hu 1993). In this study, we obtained six DH F3 and 17 DH BC1F1 lines of triticale with maintained translocations of the M-genome from Ae. ovata. Considering the homogeneity of DH plants, we transferred the Aegilops chromatin into triticale chromosomes using gametocidal gene expression, which induced chromosome rearrangements. Furthermore, we used in vitro anther cultures to generate haploids, which were developed into double haploid plants with two identical copies of triticale chromosomes with Aegilops chromatin segments (Fig. 3). The presented modus operandi showed that the Gc mechanism can be exploited in the analysis of the biology, structure and transmission of cereal chromosomes. Moreover, the Gc chromosomes can also be used in breeding as vectors for the preferential transmission of desirable traits or as inducers for chromosome aberrations.
References
Apolinarska B (1993) Stabilization of ploidy and fertility level of tetraploid triticale obtained from four cross combinations. Genet Pol 2:121–1311
Apolinarska B, Wiśniewska H, Wojciechowska B (2010) Aegilops-rye amphiploids and substitution rye used for introgression of genetic material into rye (Secale cereale L.). J Appl Genet 51:413–420
Chu CC, Wang CC, Sun CS, Hsu C, Yin KC, Chu CY, Bi FY (1975) Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen sources. Sci Sinica 18:659–668
Cuadrado A, Jouve N (1994) Mapping and organization of highly-repeated DNA sequences by means of simultaneous and sequential FISH and C-banding in 6×-triticale. Chromosome Res 2:331–338
Endo TR (1978) On the Aegilops chromosomes having gametocidal action on common wheat. In: Ramanujam S (ed) Proceedings of the 5th International Wheat Genetics Symposium, New Delhi, India, February 1978, vol 1, pp 306–314
Endo TR (1990) Gametocidal chromosomes and their induction of chromosome mutations in wheat. Jpn J Genet 65:135–152
Endo TR (2007) The gametocidal chromosome as a tool for chromosome manipulation in wheat. Chromosome Res 15:67–75
Finch RA, Miller TE, Bennett MD (1984) “Cuckoo” Aegilops addition chromosome in wheat ensures its transmission by causing chromosome breaks in meiospores lacking it. Chromosoma 90:84–88
Friebe BR, Tuleen NA, Gill BS (1999) Development and identification of a complete set of Triticum aestivum–Aegilops geniculata chromosome addition lines. Genome 42:374–380
Friebe B, Qi LL, Nasuda S, Zhang P, Tuleen NA, Gill BS (2000) Development of a complete set of Triticum aestivum–Aegilops speltoides chromosome addition lines. Theor Appl Genet 101:51–58
Friebe B, Zhang P, Nasuda S, Gill BS (2003) Characterization of a knock-out mutation at the Gc2 locus in wheat. Chromosoma 111:509–517
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res 11:4851–4865
Gradzielewska A, Gruszecka D, Leśniowska-Nowak J, Paczos-Grzęda E (2012) Identification of hybrids between triticale and Aegilops juvenalis (Thell.) Eig and determination of genetic similarity with ISSRs. Genet Mol Res 11(3):2147–2155
Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G (2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439:749–752
Gruszecka D, Tarkowski C, Stefanowska G, Marciniak K (1996) Cytological analysis of F1 Aegilops hybrids with Triticosecale. In: Guedes-Pinto H, Darvey N, Carnide VP (eds) Triticale: today and tomorrow. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 195–202
Heckmann S, Jankowska M, Schubert V, Kumke K, Ma W, Houben A (2014) Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat Commun 5:4979
Heslop-Harrison JS (2000) Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell 12:617–636
Ishihara A, Mizuno N, Islam RAKM, Doležel J, Endo TR, Nasuda S (2014) Dissection of barley chromosomes 1H and 6H by the gametocidal system. Genes Genet Syst 89:203–214
Kihara H (1954) Considerations on the evolution and distribution of Aegilops species based on the analyser-method. Cytologia 19:336–357
Knight E, Binnie A, Draeger T, Moscou M, Rey MD, Sucher J, Mehra S, King I, Moore G (2015) Mapping the ‘breaker’ element of the gametocidal locus proximal to a block of sub-telomeric heterochromatin on the long arm of chromosome 4Ssh of Aegilops sharonensis. Theor Appl Genet 128(6):1049–1059
Krowlow KD (1970) Untersuchungen über die Kreuzbarkeit zwischen Weizen und Roggen. Z Pflanzenzüchtg 64:44–72
Książczyk T, Apolinarska B, Kulak-Książczyk S, Wiśniewska H, Stojałowski S, Łapiński M (2011) Identification of the chromosome complement and the spontaneous 1R/1V translocations in allotetraploid Secale cereale × Dasypyrum villosum hybrids through cytogenetic approaches. J Appl Genet 52:305–311
Kwiatek M, Błaszczyk L, Wiśniewska H, Apolinarska B (2012) Aegilops–Secale amphiploids: chromosome categorisation, pollen viability and identification of fungal disease resistance genes. J Appl Genet 53:37–40
Kwiatek M, Wiśniewska H, Apolinarska B (2013) Cytogenetic analysis of Aegilops chromosomes, potentially usable in triticale (× Triticosecale Witt.) breeding. J Appl Genet 54:147–155
Kwiatek M, Belter J, Majka M, Wiśniewska H (2015a) Allocation of the S-genome chromosomes of Aegilops variabilis Eig. carrying powdery mildew resistance in triticale (× Triticosecale Wittmack). Protoplasma. doi:10.1007/s00709-015-0813-6
Kwiatek M, Majka M, Wiśniewska H, Apolinarska B, Belter J (2015b) Effective transfer of chromosomes carrying leaf rust resistance genes from Aegilops tauschii Coss. into hexaploid triticale (× Triticosecale Witt.) using Ae. tauschii × Secale cereale amphiploid forms. J Appl Genet 56:163–168
Kynast RG, Friebe B, Gill BS (2000) Fate of multicentric and ring chromosomes induced by a new gametocidal factor located on chromosome 4Mg of Aegilops geniculata. Chromosome Res 8:133–139
Lukaszewski AJ, Kopecký D (2010) The Ph1 locus from wheat controls meiotic chromosome pairing in autotetraploid Rye (Secale cereale L.). Cytogenet Genome Res 129:117–123
Lukaszewski AJ, Apolinarska B, Gustafson JP, Krolow KD (1984) Chromosome constitution of tetraploid triticale. Z Pflanzenzüchtg 93:222–236
Nagaki K, Tsujimoto H, Isono K, Sasakuma T (1995) Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome 38:479–486
Nasuda S, Friebe B, Gill BS (1998) Gametocidal genes induce chromosome breakage in the interphase prior to the first mitotic cell division of the male gametophyte in wheat. Genetics 149:1115–1124
Ponitka A, Ślusarkiewicz-Jarzina A, Wędzony M, Marcińska I, Woźna J (1999) The influence of various in vitro culture conditions on androgenetic embryo induction and plant regeneration from hexaploid triticale (× Triticosecale Wittm.). J Appl Genet 40:165–174
Riley R, Chapman V (1958) Genetic control of the cytological diploid behaviour of hexaploid wheat. Nature 182:713–715
Schneider A, Linc G, Molnár-Láng M, Graner A (2003) Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed 122:396–400
Schneider A, Linc G, Molnár I, Molnár-Láng M (2005) Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat–Aegilops biuncialis disomic addition lines. Genome 48:1070–1082
Schneider A, Molnár I, Molnár-Láng M (2008) Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163:1–19
Ślusarkiewicz-Jarzina A, Ponitka A (1997) Effect of genotype and media composition on embryoid induction and plant regeneration from anther culture in triticale. J Appl Genet 38:253–258
Ślusarkiewicz-Jarzina A, Ponitka A (2003) Efficient production of spontaneous and induced doubled haploid triticale plants derived from anther culture. Cereal Res Commun 31:289–296
Tsujimoto H (2005) Gametocidal genes in wheat as the inducer of chromosome breakage. In: Tsunewaki K (ed) Frontiers of wheat bioscience. Memorial issue, Wheat Information Service No. 100. Kihara Memorial Yokohama Foundation, pp 33–48
Tsujimoto H, Tsunewaki K (1985) Gametocidal genes in wheat and its relatives. II. Suppressor of the chromosome 3C gametocidal gene of Aegilops triuncialis. Can J Genet Cytol 27:178–185
Unfried I, Gruendler P (1990) Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res 18:4011
van Slageren MW (1994) Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae). Agricultural University of Wageningen, the Netherlands, and International Center for Agricultural Research in Dry Areas, Aleppo, Syria
Vershinin A, Svitashev S, Gummesson P-O, Salomon B, von Bothmer R, Bryngelsson T (1994) Characterization of a family of tandemly repeated DNA sequences in Triticeae. Theor Appl Genet 89:217–225
Vrána J, Kubaláková M, Simková H, Cíhalíková J, Lysák MA, Dolezel J (2000) Flow sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 156:2033–2041
Wang P, Chen Y (1983) Preliminary study on production of height of pollen H2 generation in winter wheat grown in the field. Acta Agron Sin 9:283–284
Wang YB, Hu H (1993) Gamete composition and chromosome variation in pollen-derived plants from octoploid triticale × common wheat hybrids. Theor Appl Genet 85:681–687
Wiśniewska H, Kwiatek M, Kulak-Książczyk S, Apolinarska B (2013) Introgression of A- and B-genome of tetraploid triticale chromatin into tetraploid rye. J Appl Genet 54:435–440
Wojciechowska B, Pudelska H (2002) Hybrids and amphiploids of Aegilops ovata L. with Secale cereale L.: production, morphology and fertility. J Appl Genet 43:415–421
Zhuang JJ, Xu J (1983) Increasing differentiation frequencies in wheat pollen callus. In: Cell and tissue culture techniques for cereal crop improvement. Science Press, Beijing, p 431
Zwierzykowski Z, Zwierzykowska E, Taciak M, Jones N, Kosmala A, Krajewski P (2008) Chromosome pairing in allotetraploid hybrids of Festuca pratensis × Lolium perenne revealed by genomic in situ hybridization (GISH). Chromosome Res 16:575–585
Acknowledgements
This work was financed by the National Science Centre (DEC-2013/11/D/NZ9/02719). We would like to gratefully acknowledge the technical assistance of Mrs. Grażyna Cicha and Mrs. Joanna Maszner. We also thank Prof. Barbara Apolinarska for her priceless suggestions and clues.
Author contributions
M. Kwiatek initiated the project. M. Kwiatek and H. Wiśniewska designed the study. M. Kwiatek and J. Belter obtained the hybrid plants. A. Ślusarkiewicz-Jarzina, A. Ponitka and H. Pudelska produced the double haploid lines. M. Kwiatek and M. Majka performed the GISH/FISH analysis. M. Kwiatek wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Andrzej Górny
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kwiatek, M., Majka, M., Ślusarkiewicz-Jarzina, A. et al. Transmission of the Aegilops ovata chromosomes carrying gametocidal factors in hexaploid triticale (×Triticosecale Wittm.) hybrids. J Appl Genetics 57, 305–315 (2016). https://doi.org/10.1007/s13353-015-0332-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-015-0332-3