Abstract
Anthracnose caused by Colletotrichum gloeosporioides is an economic disease that affects soybean production worldwide. This study developed a rapid, sensitive method for the detection of C. gloeosporioides using a loop-mediated isothermal amplification (LAMP) assay. By targeting a glutamine synthetase (GS) gene sequence, the GS-Cg-LAMP assay works most efficiently at 64°C and allows the detection of C. gloeosporioides DNA within 70 min based on a color change from orange to yellow-green following the addition of SYBR Green I to the LAMP reaction products. The detection limit was 1 pg μL−1 of genomic DNA per reaction. In specificity tests, the positive reaction (yellow-green color by the naked eye) was observed only in the presence of C. gloeosporioides, and none of other Colletotrichum spp. or fungal isolates produced a color change. Moreover, the GS-Cg-LAMP assay was successfully used to diagnose anthracnose caused by C. gloeosporioides in diseased soybean samples obtained from the field and detected the pathogen in soybean seeds bought from farmers’ markets. Our study provides a simple, sensitive tool for the rapid diagnosis of soybean anthracnose caused by C. gloeosporioides.
Similar content being viewed by others
Introduction
Colletotrichum gloeosporioides is an economically important pathogen that infects a wide range of crops causing enormous production losses each year (Jeffries 1990). The fungus causes anthracnose disease of cereals, legumes, vegetables, perennial crops, and tree fruits (Phoulivong et al. 2010). As a seed-borne disease (Manandhar et al. 1987), anthracnose caused by C. gloeosporioides damages soybeans commencing at the seedling stage up to harvest (Kwon et al. 2013), and the disease can affect all of the above ground parts of the plant. Irregularly shaped, brown lesions (Sugawara et al. 2009), which are the most distinctive symptom, appear on stems, leaves, and pods during the early reproductive stage. Seeds and crop residues are considered to be the primary sources of the infection (Chen et al. 2006).
In addition to Colletotrichum gloeosporioides, C. truncatum, C. capsici, C. dematium, C. destructivum and C. coccodes also cause soybean anthracnose (Ghosh et al. 2016; Lou et al. 2009; Hartman et al. 1988; Roy 1982). These species are difficult to differentiate by microscopic examination because of their morphological similarity. Moreover, the disease symptoms on soybean plants caused by the Colletotrichum spp. listed above and C. gloeosporioides are similar and difficult to differentiate. Therefore, new techniques for the rapid diagnosis of anthracnose caused by C. gloeosporioides are needed.
In recent years, numerous molecular methods have been developed to detect pathogens. The most common molecular detection method is based on the polymerase chain reaction (PCR). A PCR assay has been reported for the detection of C. gloeosporioides, but it has intrinsic disadvantages, including low amplification efficiency and a complicated experimental process. Loop-mediated isothermal amplification (LAMP) (Tsugunori et al. 2000) of DNA, an alternative technology, can be applied for the diagnosis of viruses (Przewodowska et al. 2015), bacteria (Wen et al. 2011), and fungi (Sun et al. 2010). This assay requires a set of six primers and the large fragment of Bst DNA polymerase to amplify DNA quickly under isothermal conditions. The LAMP products are judged with the naked eye after the addition of SYBR Green I (Karlsen et al. 1995). Although this method has been developed for the detection of some soybean pathogens, such as Fusarium equiseti (Lu et al. 2015), F. asiaticum (Xu et al. 2017), and Colletotrichum truncatum (Tian et al. 2016), no method for detecting C. gloeosporioides has been reported.
In this study, we used the glutamine synthetase (GS) gene as the target sequence to develop a rapid, sensitive LAMP assay for the detection of C. gloeosporioides. The assay can be used for the rapid diagnosis of anthracnose disease caused by C. gloeosporioides by directly detecting the pathogen in diseased soybean tissues in the field.
Materials and methods
Source of strains
Colletotrichum gloeosporioides strains were obtained from diseased leaves, stems, and pods of soybean plants collected in the Shandong, Jiangsu, Anhui, and Hebei Provinces, China. The strains of C. gloeosporioides, C. truncatum, and C. acutatum were identified morphologically and by internal transcribed spacer (ITS) sequencing before use and are maintained in our laboratory. Table 1 lists the fungal species and isolates used in this study.
Culture conditions and DNA extraction from pure cultures
C. gloeosporioides and other fungi were cultured in potato dextrose agar (PDA) (200 g potato extract L−1, 2% [w/v] glucose, and 2% [w/v] agar, autoclaved at 120 °C for 20 min). Mycelia of each isolate were obtained by growing the strains in potato dextrose broth (200 g potato extract L−1, 2% [w/v] glucose, autoclaved at 120 °C for 20 min) at 25 °C for 5 days. Phytophthora sojae was cultured on V8 medium and P. sojae mycelia were cultured in V8 juice broth. Mycelium DNA was extracted according to the CTAB protocol described by Möller et al. (Möller et al. 1992). DNA concentrations were quantified using a NanoDrop spectrophotometer ND-1000 (Thermo Fisher Scientific) and stored at −20 °C.
LAMP primer design and screening
Selection of a suitable target for LAMP primers is critical. After comparing different target gene sequences among strains of C. gloeosporioides and similar species at the National Center for Biotechnology Information website, the glutamine synthetase (GS) gene (GenBank ID: DQ792872.1) was chosen as the target DNA sequence. Glutamine synthetase is used to control nitrogen metabolism. The GS gene is present in fungi, plants, and animals. We compared the GS sequences of C. gloeosporioides with those of other Colletotrichum spp., and used a sequence specific to C. gloeosporioides (Fig. 1) to design LAMP primers. The LAMP primers were designed using PrimerExplorer V4 software (http://primerexplorer.jp/e/). Multiple sets of primers were designed and screened using a series of specificity and sensitivity tests. Finally, a set of four primers with high species specificity and sensitivity, targeting the GS sequence of C. gloeosporioides, was selected for further study. The loop primer (LF and LB) was used to accelerate the reaction speed. The primer sequences are shown in Table 2.
LAMP reaction and product detection
The LAMP assay was performed in a reaction volume of 25 μL, which contained 0.8 μM each of FIP and BIP, 0.1 μM each of F3, B3, LF, and LB, 1.4 mM dNTPs, 0.8 MBetaine, 0.8 M Tris-HCl (pH 8.8), 0.4 mM KCl, 0.4 mM (NH4)2SO4, 4% Triton X-100, 8 U Bst DNA polymerase, and 4 μL of DNA sample. The amplification reaction was run at 64 °C for 70 min. Each lamp assay included positive (a sample with C. gloeosporioides DNA) and negative (a sample to which no template was added) controls and each sample was analyzed at least three times.
After the reaction, a LAMP product was sought directly by the unaided eye after adding 0.25 μL of SYBR Green I (10,000× concentrate in dimethyl sulfoxide; Life Technologies, Carlsbad, CA, USA). With a positive reaction, a yellow-green color was clearly observed with the naked eye, whereas the orange color remained with a negative reaction.
DNA extraction from diseased soybean tissues
Soybean seeds were planted in vermiculite and grown in a greenhouse at 25 °C. Plugs of agar (5X5) with C. gloeosporioides mycelia were inoculated on soybean leaves in vitro and incubated at 25 °C for 5 days (24 h in the dark, followed by a 12 h photoperiod). DNA was extracted from the diseased leaves using the DNAsecure Plant Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. As a control, the same method was used to extract DNA from non-inoculated healthy soybean leaves.
To clarify the feasibility of the LAMP assay for the rapid diagnosis of anthracnose caused by C. gloeosporioides, suspected diseased soybean samples were collected from the field. DNA from the tissues of diseased leaves, pods, and stems was extracted in the laboratory using a DNAsecure plant kit (Tiangen) according to the manufacturer’s protocol. All samples were stored at −20 °C.
DNA extraction from seeds containing C. gloeosporioides conidia
Ten soybean seed samples were purchased from different farmers’ markets. To detect C. gloeosporioides in the samples, 50 g of seeds from each sample were soaked and shocked with 100 mL of deionized water and 3–5 drops of 20% (v/v) Tween in 250-mL Erlenmeyer flasks for 30 min. The mixture was filtered through a sieve and the filtrate collected and centrifuged at 6500 rpm for 10 min. The DNA was extracted from the precipitate using a Power Soil® DNA Isolation Kit (MOBIO) according to the manufacturer’s protocol. The DNA was stored at −20 °C.
Assays were also performed by adding conidia of C. gloeosporioides to soybean seed samples that were negative by LAMP detection to determine the detection limit of the GS-Cg-LAMP assay. Spore suspensions of C. gloeosporioides containing various numbers of conidia (0, 10, 50, 100, 1000, and 10,000) were added to 50 g of soybean seeds. DNA was extracted using the method described above and stored at −20 °C.
Results
Specificity of the GS-cg-LAMP assay
The strains of C. gloeosporioides isolated from different hosts and different areas, several other Colletotrichum spp., and isolates of other fungi were used to confirm the specificity of the GS-Cg-LAMP assay. As shown in Fig. 2 and Table 1, only the samples containing C. gloeosporioides gave positive reactions as indicated by a visible yellow-green color, whereas reactions containing other Colletotrichum spp. or other fungi isolates were negative (orange color). Assays were performed using serial ten-fold dilutions (ranging from 100 ng to 1 fg) of pure C. gloeosporioides genomic DNA to determine the detection limit of the GS-Cg-LAMP assay. The minimum concentration of C. gloeosporioides DNA detected with the GS-Cg-LAMP assay was 1 pg μL−1. This indicated that the GS-Cg-LAMP assay can be used to detect C. gloeosporioides specifically.
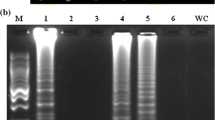
Generality of the GS-Cg-LAMP assay for different Colletotrichum gloeosporioides isolates. The C. gloeosporioides isolated from different hosts and different areas were showed as follows: 1, soybean of Shandong; 2, soybean of Jiangsu; 3, soybean of Sichuan; 4, peanut of Hubei; 5–6, grape of Jiangsu; 7–8, pear of Jiangsu; 9–12, rubber of Hainan; 13, C. fishtail palm of Hainan; 14, navel orange of Hainan; 15, negative control
Detection of C. gloeosporioides in diseased soybean tissues
We extracted the DNA from diseased soybean leaves inoculated with C. gloeosporioides to simulate field conditions. All the inoculated soybean tissues reacted positively, similar to the positive control.
We next used the GS-Cg-LAMP assay to detect C. gloeosporioides in samples suspected of having soybean anthracnose collected from the field in the Jiangsu, Anhui, and Hubei Provinces. Of 152 samples analyzed, 65 tested positive in the LAMP assay. Of these samples, 22 were identified using traditional isolation methods (Table 3). The results show that our GS-Cg-LAMP assay was both rapid and accurate compared with traditional isolation methods. The identification of isolates based on morphological characteristics and ITS sequence alignment was consistent with the assay results. The GS-Cg-LAMP assay reported here can be used to diagnose soybean anthracnose caused by C. gloeosporioides in the field rapidly.
Detection of C. gloeosporioides in soybean seeds bought from farmers’ markets
The GS-Cg-LAMP assay was used to detect C. gloeosporioides in ten samples of soybean seeds obtained from farmers’ markets. C. gloeosporioides was identified in four samples from the Jiangsu, Shandong, and Zhejiang Provinces.
To evaluate the efficiency of the LAMP assay for detecting C. gloeosporioides in soybean seeds, conidia of C. gloeosporioides were added to pathogen-free samples of soybean seeds manually. The GS-Cg-LAMP assay was positive when there were ten or more conidia in 50 g of soybean seeds.
Discussion
This study developed an effective method for detecting C. gloeosporioides from diseased soybean samples and rapidly diagnosing soybean anthracnose caused by the pathogen in the field. The GS-Cg-LAMP assay can also detect C. gloeosporioides in soybean seeds efficiently.
The LAMP system that we developed for the detection of C. gloeosporioides uses four specific primers and two loop primers, the designs of which were based on the sequence of the glutamine synthetase gene (GS), which is common to fungi, plants, and animals. A blast search indicated that the GS gene sequence was conserved in different C. gloeosporioides strains but had rich polymorphisms among related Colletotrichum spp. Consequently, the GS gene has the potential to be used as a species-specific target to detect C. gloeosporioides.
A PCR-based detection assay for C. gloeosporioides using a forward primer (CgInt) from a conserved rDNA region and a universal primer (ITS4) has been reported. However, false-positive reactions can still occur because the ITS sequence is highly conserved in closely related species. The attempted detection of pathogens in samples may produce erroneous results due to nonspecific binding of the primer pairs. Moreover, the amplified PCR products must be separated using gel electrophoresis, strained with ethidium bromide, and examined with ultraviolet light, which requires at least six hours to complete.
Compared with PCR and traditional methods of detecting C. gloeosporioides in diseased soybean tissues, the GS-CG-LAMP assay has several advantages. First, high specificity is afforded by the use of four primers that recognize six distinct regions of the template DNA, unlike the PCR method, which uses only two primers defining two regions of the template. Second, the GS-Cg-LAMP assay is simple to perform, with only 2 h needed to detect the pathogen, and the results can be visualized with the unaided eye. Third, the GS-Cg-LAMP assay is very efficient, as C. gloeosporioides can be detected directly and specifically in a mixture containing the DNA of the host plant, C. gloeosporioides, and saprophytic microbes. The assay can be used directly on samples obtained from diseased soybean tissues from the field or soybean seeds purchased in markets.
In conclusion, we developed a LAMP assay for the specific detection of C. gloeosporioides in diseased soybean tissues, allowing the rapid diagnosis of soybean anthracnose caused by the pathogen. We also used the assay to detect C. gloeosporioides strains from other diseased hosts, including grapes, pears, and rubber, and all strains produced positive reactions with the GS-Cg-LAMP assay.
References
Chen LS, Chu C, Liu CD, Chen RS, Tsay JG (2006) PCR-based detection and differentiation of anthracnose pathogens, Colletotrichum gloeosporioides and C. truncatum, from vegetable soybean in Taiwan. J Phytopathol 154(11–12):654–662. https://doi.org/10.1111/j.1439-0434.2006.01163.x
Ghosh R, Bhadra S, Bandyopadhyay M (2016) Morphological and molecular characterization of Colletotrichum capsici causing leaf-spot of soybean. Tropical plant research 3(3):481–490. https://doi.org/10.22271/tpr.2016.v3.i3.064
Hartman GL, Manandhar JB, Sinclair JB (1988) Incidence of Colletotrichum dematium on prickly sida, spotted spurge and smooth pigweed and pathogenicity to soybean. Plant Dis 72(72):390–393
Jeffries P, Dodd JC, Jeger MJ, Plumbley RA (1990) The biology and control of colletotrichum species on tropical fruit crops. MJ and Plumbley 39(3):343–366
Karlsen F, Steen HB, Nesland JM (1995) SYBR green I DNA staining increases the detection sensitivity of viruses by polymerase chain reaction. J Virol Methods 55(1):153
Kwon J-H, Kim J, Choi O, Gang G-H, Han S, Kwak Y-S (2013) Anthracnose caused by Colletotrichum horiion sweet persimmon in Korea: dissemination of conidia and disease development. J Phytopathol 161(7–8):497–502. https://doi.org/10.1111/jph.12096
Lou BG, Chen WJ, Lin C, Wang GR, Xia GM, Lou MQ (2009) A new symptom type of soybean pod anthracnose and identification of its pathogen. Acta Phytophylacica Sinica 36:229–233
Lu C, Zhang H, Wang Y, Zheng X (2015) Rapid diagnosis of fusarium root rot in soybean caused by Fusarium equiseti or Fusarium graminearum using loop-mediated isothermal amplification (LAMP) assays. Australas Plant Pathol 44(4):437–443. https://doi.org/10.1007/s13313-015-0361-8
Manandhar JB, Thapliyal PN, Cavanaugh KJ, Sinclair JB (1987) Manandhar et al. 1987.pdf. Mycopathologia 98(2):69–75
Möller EM, Bahnweg G, Sandermann H, Geiger HH (1992) A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res 20(22):6115–6116
Phoulivong S, Cai L, Chen H, McKenzie EHC, Abdelsalam K, Chukeatirote E, Hyde KD (2010) Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Divers 44(1):33–43. https://doi.org/10.1007/s13225-010-0046-0
Przewodowska A, Zacharzewska B, Chołuj J, Treder K (2015) A one-step, real-time reverse transcription loop mediated isothermal amplification assay to detect potato virus Y. Am J Potato Res 92(3):303–311. https://doi.org/10.1007/s12230-015-9430-3
Roy KW (1982) Seedling diseases caused in soybean by species of Colletotrichum and Glomerella. Phytopathology 72:1093–1096
Sugawara K, Matsudate A, Ito Y, Namai T (2009) Anthracnose of Christmas rose caused by Colletotrichum sp. J Gen Plant Pathol 75(2):163–166. https://doi.org/10.1007/s10327-009-0151-7
Sun J, Najafzadeh MJ, Vicente V, Xi L, de Hoog GS (2010) Rapid detection of pathogenic fungi using loop-mediated isothermal amplification, exemplified by Fonsecaea agents of chromoblastomycosis. J Microbiol Methods 80(1):19–24. https://doi.org/10.1016/j.mimet.2009.10.002
Tian Q, Lu C, Wang S, Xiong Q, Zhang H, Wang Y, Zheng X (2016) Rapid diagnosis of soybean anthracnose caused by Colletotrichum truncatum using a loop-mediated isothermal amplification (LAMP) assay. Eur J Plant Pathol 148(4):785–793. https://doi.org/10.1007/s10658-016-1132-2
Tsugunori N, Okayama H, Mea H (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12):E63
Wen P, Jia-Ying W, Hai-Yan S et al (2011) Development and application of the novel visual loop-mediated isothermal amplification of Omp25 sequence for rapid detection of Brucella sp. Journal of Animal & Veterinary Advances 10(16):2120–2126
Xu M, Ye W, Zeng D, Wang Y, Zheng X (2017) Rapid diagnosis of wheat head blight caused by Fusarium asiaticum using a loop-mediated isothermal amplification assay. Australas Plant Pathol 46(3):261–266. https://doi.org/10.1007/s13313-017-0487-y
Acknowledgements
This research was supported by the National High-Tech R&D Program (863 Program) (2012AA101501), the National Department Public Benefit Research Foundation (200903004), Public Sector Research Funding (201303018).
Author information
Authors and Affiliations
Corresponding author
Additional information
The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see:
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, S., Ye, W., Tian, Q. et al. Rapid detection of Colletotrichum gloeosporioides using a loop-mediated isothermal amplification assay. Australasian Plant Pathol. 46, 493–498 (2017). https://doi.org/10.1007/s13313-017-0511-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-017-0511-2