Abstract
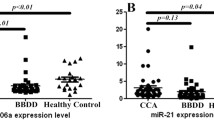
Altered microRNA (miRNA) expression plays a role in cholangiocarcinoma (CCA) development; thus, detection of blood-circulating miRNAs could be useful as CCA markers. This study profiled serum miRNA levels in patients with primary sclerosing cholangitis (PSC) and CCA and then assessed the role of miR-150-5p in CCA progression in vitro. Three samples were randomly selected from each of 50 sera of healthy controls, 30 PSC sera, and 28 CCA sera with matched bile samples for miRNA microarray profiling. The dysregulated miRNAs were confirmed using qRT-PCR, and miR-150-5p was selected for further in vitro and ex vivo studies. The miRNA microarray identified three dysregulated miRNAs in both CCA and PSC samples, while miR-150-5p level was consistently lower in CCA sera, bile, and tissues than in normal control and PSC sera (P < 0.05). Furthermore, levels of miR-150-5p were associated with serum carbohydrate antigen 19-9 (CA19-9) levels and CCA pathological grade. Bioinformatic Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses showed that miR-150-5p could regulate hand-full gene pathways, including cancer pathway (P < 0.01). However, overexpression of miR-150-5p inhibited proliferation, migration, and invasion capability of CCA cells (P < 0.05). Luciferase reporter assay showed that miR-150-5p bound to an oncogene Ets including gene-1 (ELK1), and Western blot data confirmed that miR-150-5p suppressed ELK1 expression in CCA cell lines. These results suggest that reduced miR-150-5p expression could contribute to CCA development and progression due to uncontrolled ELK1 expression. Thus, further study could evaluate miR-150-5p as a novel target and predictor for CCA prevention and treatment.





Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004;24(2):115–25. doi:10.1055/s-2004-828889.
Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366(9493):1303–14. doi:10.1016/S0140-6736(05)67530-7.
Malhi H, Gores GJ. Cholangiocarcinoma: modern advances in understanding a deadly old disease. J Hepatol. 2006;45(6):856–67. doi:10.1016/j.jhep.2006.09.001.
Rizvi S, Eaton JE, Gores GJ. Primary sclerosing cholangitis as a premalignant biliary tract disease: surveillance and management. Clin Gastroenterol Hepatol: Off Clin Pract J Am Gastroenterol Assoc. 2015;13(12):2152–65. doi:10.1016/j.cgh.2015.05.035.
Charatcharoenwitthaya P, Lindor KD. Primary sclerosing cholangitis: diagnosis and management. Curr Gastroenterol Rep. 2006;8(1):75–82.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153(3):516–9. doi:10.1016/j.cell.2013.04.003.
Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2014. doi:10.1016/j.addr.2014.09.001.
Silakit R, Loilome W, Yongvanit P, Chusorn P, Techasen A, Boonmars T, et al. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: a prospective prognostic indicator. J Hepato-Biliary-Pancreat Sci. 2014;21(12):864–72. doi:10.1002/jhbp.145.
Li L, Masica D, Ishida M, Tomuleasa C, Umegaki S, Kalloo AN, et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014;60(3):896–907. doi:10.1002/hep.27050.
Joung JG, Hwang KB, Nam JW, Kim SJ, Zhang BT. Discovery of microRNA-mRNA modules via population-based probabilistic learning. Bioinformatics. 2007;23(9):1141–7. doi:10.1093/bioinformatics/btm045.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Gene Ontol Consortium Nat Genet. 2000;25(1):25–9. doi:10.1038/75556.
Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–45. doi:10.1101/gr.6202607.
An F, Gong B, Wang H, Yu D, Zhao G, Lin L, et al. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis: Int J Program Cell Death. 2012;17(7):702–16. doi:10.1007/s10495-012-0704-7.
An F, Zhan Q, Xia M, Jiang L, Lu G, Huang M, et al. From moderately severe to severe hypertriglyceridemia induced acute pancreatitis: circulating MiRNAs play role as potential biomarkers. PLoS One. 2014;9(11):e111058. doi:10.1371/journal.pone.0111058.
Eaton JE, Talwalkar JA, Lazaridis KN, Gores GJ, Lindor KD. Pathogenesis of primary sclerosing cholangitis and advances in diagnosis and management. Gastroenterology. 2013;145(3):521–36. doi:10.1053/j.gastro.2013.06.052.
Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145(6):1215–29. doi:10.1053/j.gastro.2013.10.013.
Patel T. Worldwide trends in mortality from biliary tract malignancies. BMC Cancer. 2002;2:10.
Palmer WC, Patel T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J Hepatol. 2012;57(1):69–76. doi:10.1016/j.jhep.2012.02.022.
Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–56. doi:10.1038/nrclinonc.2014.5.
Voigtlander T, Gupta SK, Thum S, Fendrich J, Manns MP, Lankisch TO, et al. MicroRNAs in serum and bile of patients with primary sclerosing cholangitis and/or cholangiocarcinoma. PLoS One. 2015;10(10):e0139305. doi:10.1371/journal.pone.0139305.
Bernuzzi F, Marabita F, Lleo A, Carbone M, Mirolo M, Marzioni M, et al. Serum micrornas as novel biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Clin Exp Immunol. 2016. doi:10.1111/cei.12776.
Letelier P, Riquelme I, Hernandez AH, Guzman N, Farias JG, Roa JC. Circulating microRNAs as biomarkers in biliary tract cancers. Int J Mol Sci. 2016;17(5):E791. doi:10.3390/ijms17050791.
Razumilava N, Gores GJ, Lindor KD. Cancer surveillance in patients with primary sclerosing cholangitis. Hepatology. 2011;54(5):1842–52. doi:10.1002/hep.24570.
Honda N, Jinnin M, Kira-Etoh T, Makino K, Kajihara I, Makino T, et al. miR-150 down-regulation contributes to the constitutive type I collagen overexpression in scleroderma dermal fibroblasts via the induction of integrin beta3. Am J Pathol. 2013;182(1):206–16. doi:10.1016/j.ajpath.2012.09.023.
Patel AH, Harnois DM, Klee GG, LaRusso NF, Gores GJ. The utility of CA 19-9 in the diagnoses of cholangiocarcinoma in patients without primary sclerosing cholangitis. Am J Gastroenterol. 2000;95(1):204–7. doi:10.1111/j.1572-0241.2000.01685.x.
Vasilatou D, Papageorgiou S, Pappa V, Papageorgiou E, Dervenoulas J. The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol. 2010;84(1):1–16. doi:10.1111/j.1600-0609.2009.01348.x.
Feng J, Yang Y, Zhang P, Wang F, Ma Y, Qin H, et al. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-Myb. J Cell Mol Med. 2014;18(10):2125–34. doi:10.1111/jcmm.12398.
Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, et al. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32(12):1832–9. doi:10.1093/carcin/bgr223.
Wang WH, Chen J, Zhao F, Zhang BR, Yu HS, Jin HY, et al. MiR-150-5p suppresses colorectal cancer cell migration and invasion through targeting MUC4. Asian Pac J Cancer Prev: APJCP. 2014;15(15):6269–73.
Ito M, Teshima K, Ikeda S, Kitadate A, Watanabe A, Nara M, et al. MicroRNA-150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6, in advanced cutaneous T-cell lymphoma. Blood. 2014;123(10):1499–511. doi:10.1182/blood-2013-09-527739.
Sakr M, Takino T, Sabit H, Nakada M, Li Z, Sato H. miR-150-5p and miR-133a suppress glioma cell proliferation and migration through targeting membrane-type-1 matrix metalloproteinase. Gene. 2016;587(2):155–62. doi:10.1016/j.gene.2016.04.058.
Assumpcao MB, Moreira FC, Hamoy IG, Magalhaes L, Vidal A, Pereira A, et al. High-throughput miRNA sequencing reveals a field effect in gastric cancer and suggests an epigenetic network mechanism. Bioinforma Biol Insights. 2015;9:111–7. doi:10.4137/BBI.S24066.
Chai Y, Chipitsyna G, Cui J, Liao B, Liu S, Aysola K, et al. c-Fos oncogene regulator Elk-1 interacts with BRCA1 splice variants BRCA1a/1b and enhances BRCA1a/1b-mediated growth suppression in breast cancer cells. Oncogene. 2001;20(11):1357–67. doi:10.1038/sj.onc.1204256.
Hsieh YH, TT W, Tsai JH, Huang CY, Hsieh YS, Liu JY. PKCalpha expression regulated by Elk-1 and MZF-1 in human HCC cells. Biochem Biophys Res Commun. 2006;339(1):217–25. doi:10.1016/j.bbrc.2005.11.015.
Acknowledgments
The authors would like to thank our staff in both Departments for their contributions to this study and Genminix Company for their technical assistance. This study was supported in part by grants from the National Natural Science Foundation of China (no. 81502038 to FA, no. 81302382 to YJ, and no. 31400720 to WW), the Natural Science Foundation of Jiangsu Province (no. BK2012098 to FA), and the Wuxi Municipal Science and Technology Development Planning Funds (no. CSZ00N1304 to FA).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This study was approved by the Institutional Review Board (IRB) of Nanjing Medical University. All patients provided written informed consent before participation of this study.
Conflicts of interest
None
Additional information
Xiongbo Wu and Min Xia are authors contributed equally to this work.
Electronic supplementary material
Table S1
(Clinical characteristics of CCA patients. The table shows the relevant clinical information of CCA patients. The gender, age, tumor location, CA-19-9 and pathological grade of 13 CCA patients with tumor tissues and matched adjacent-tumor tissues were listed. DOCX 13 kb)
Fig. S1
(The Gene Ontology (GO) analyze of 3 differentially expressed miRNAs-regulated genes in body fluids of CCAs. The GO term predicted functions of genes regulated by these 3 miRNAs (P < 0.001). The vertical axis represented GOs and the horizontal axis is enrichment degree. Enrichment degree means the contribution of one miRNA to the surrounding GOs or the contribution one GO to the surrounding miRNAs. The key functions always have the higher enrichment degrees. The top 50 enrichment of GOs were showed. JPEG 434 kb)
Fig. S2
(The Kyoto Encyclopedia of Genes and Genomes (KEGG) analyze of 3 differentially expressed miRNAs-regulated pathways in body fluids of CCAs. KEGG predicted 29 pathways related to tumors regulated by these 3 miRNAs, (P < 0.001). The vertical axis is the pathway category and the horizontal axis is the P value of each pathway. The lower P value was, the more miRNAs regulated the pathways and the pathways had more important roles in CCA. JPEG 195 kb)
Fig. S3
(miRNA-mRNA network. The miRNAs target genes were predicted by TargetScan and Sanger and then miRNA–mRNA-network was generated. The blue circles represent genes, and the blue squares represent miRNAs. The lines indicate the relationship between genes and miRNA. The degree (size) of the blue square indicates the regulatory functionality of miRNA, i.e., the greater the degree, the more functions of the miRNA possess. The figure shows the relationship among these 3 overlapping miRNAs and their targeted genes, including tumor development and miR-150-5p targeted ELK1, Notch3, and MMP14. PNG 1689 kb)
.
Fig. S4
(Assay for the transfection efficiency. 150-5pM, NSM, 150-5pI, NSI were transiently transfected into HCCC-9810 and RBE cells respectively for 24 h and then subjected to qRT-PCR analysis of miR-150-5p expression. qRT-PCR was in triplicate and repeated three times. JPEG 590 kb)
Rights and permissions
About this article
Cite this article
Wu, X., Xia, M., Chen, D. et al. Profiling of downregulated blood-circulating miR-150-5p as a novel tumor marker for cholangiocarcinoma. Tumor Biol. 37, 15019–15029 (2016). https://doi.org/10.1007/s13277-016-5313-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5313-6