Abstract
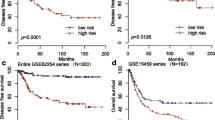
Gastric cancer (GC) is one of the most aggressive malignancies and has a poor prognosis. Identifying novel diagnostic and prognostic markers is of great importance for the management and treatment of GC. Long non-coding RNAs (lncRNAs), which are involved in multiple processes during the development and progression of cancer, may act as potential biomarkers of GC. Here, by performing data mining using four microarray data sets of GC downloaded from the Gene Expression Omnibus (GEO) database with different classifiers and risk score analyses, we identified a five-lncRNA signature (AK001094, AK024171, AK093735, BC003519 and NR_003573) displaying both diagnostic and prognostic values for GC. The results of the Kaplan-Meier survival analysis and log-rank test showed that the risk score based on this five-lncRNA signature was closely associated with overall survival time (p = 0.0001). Further analysis revealed that the risk score is an independent predictor of prognosis. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of 30 pairs of GC tissue samples confirmed that the five lncRNAs were dysregulated in GC, and receiver operating characteristic (ROC) curves showed the high diagnostic ability of combining the five lncRNAs, with an area under the curve (AUC) of 0.95 ± 0.025. The five lncRNAs involved in several cancer-related pathways were identified using gene set enrichment analysis (GSEA). These findings indicate that the five-lncRNA signature may have a good clinical applicability for determining the diagnosis and predicting the prognosis of GC.






Similar content being viewed by others
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32.
Sun Z, Wang ZN, Zhu Z, Xu YY, Xu Y, Huang BJ, et al. Evaluation of the seventh edition of American Joint Committee on Cancer TNM staging system for gastric cancer: results from a Chinese monoinstitutional study. Ann Surg Oncol. 2012;19(6):1918–27.
Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–87.
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–61.
Stower H. Epigenetics: X inactivation by titration. Nat Rev Genet. 2013;14(8):518.
Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8.
Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66(10):5330–7.
Li H, Yu B, Li J, Su L, Yan M, Zhu Z, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–29.
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92.
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282(34):24731–42.
Day JR, Jost M, Reynolds MA, Groskopf J, Rittenhouse H. PCA3: from basic molecular science to the clinical lab. Cancer Lett. 2011;301(1):1–6.
Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–10.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64.
Risueno A, Fontanillo C, Dinger ME, De Las Rivas J. GATExplorer: genomic and transcriptomic explorer; mapping expression probes to gene loci, transcripts, exons and ncRNAs. BMC Bioinformatics. 2010;11:221.
Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0—an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35(Database issue):D178–82.
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41(17):e166.
Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59(5):579–85.
Meng J, Li P, Zhang Q, Yang Z, Fu S. A four-long non-coding RNA signature in predicting breast cancer survival. J Exp Clin Cancer Res. 2014;33:84.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50.
Du J, Yuan Z, Ma Z, Song J, Xie X, Chen Y. KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol BioSyst. 2014;10(9):2441–7.
Nong Y, Wu D, Lin Y, Zhang Y, Bai L, Tang H. Tenascin-C expression is associated with poor prognosis in hepatocellular carcinoma (HCC) patients and the inflammatory cytokine TNF-alpha-induced TNC expression promotes migration in HCC cells. Am J Cancer Res. 2015;5(2):782–91.
Hao NB, Tang B, Wang GZ, Xie R, CJ H, Wang SM, et al. Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-kappaB signaling pathway for gastric cancer metastasis. Cancer Lett. 2015;361(1):57–66.
Xiao YC, Yang ZB, Cheng XS, Fang XB, Shen T, Xia CF, et al. CXCL8, overexpressed in colorectal cancer, enhances the resistance of colorectal cancer cells to anoikis. Cancer Lett. 2015;361(1):22–32.
Marra P, Mathew S, Grigoriadis A, Wu Y, Kyle-Cezar F, Watkins J, et al. IL15RA drives antagonistic mechanisms of cancer development and immune control in lymphocyte-enriched triple-negative breast cancers. Cancer Res. 2014;74(17):4908–21.
Gu J, Ding JY, CL L, Lin ZW, Chu YW, Zhao GY, et al. Overexpression of CD88 predicts poor prognosis in non-small-cell lung cancer. Lung Cancer. 2013;81(2):259–65.
Span PN, Pollakis G, Paxton WA, Sweep FC, Foekens JA, Martens JW, et al. Improved metastasis-free survival in nonadjuvantly treated postmenopausal breast cancer patients with chemokine receptor 5 del32 frameshift mutations. Int J Cancer. 2015;136(1):91–7.
Palagani V, Bozko P, El Khatib M, Belahmer H, Giese N, Sipos B, et al. Combined inhibition of Notch and JAK/STAT is superior to monotherapies and impairs pancreatic cancer progression. Carcinogenesis. 2014;35(4):859–66.
Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–46.
Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123(22):e123–33.
Oshima H, Ishikawa T, Yoshida GJ, Naoi K, Maeda Y, Naka K, et al. TNF-alpha/TNFR1 signaling promotes gastric tumorigenesis through induction of Noxo1 and Gna14 in tumor cells. Oncogene. 2014;33(29):3820–9.
Yeh TS, Wu CW, Hsu KW, Liao WJ, Yang MC, Li AF, et al. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygenase-2. Cancer Res. 2009;69(12):5039–48.
Kim YH, Liang H, Liu X, Lee JS, Cho JY, Cheong JH, et al. AMPKalpha modulation in cancer progression: multilayer integrative analysis of the whole transcriptome in Asian gastric cancer. Cancer Res. 2012;72(10):2512–21.
Onodera Y, Nam JM, Bissell MJ. Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-GlcNAc pathways. J Clin Invest. 2014;124(1):367–84.
Widau RC, Parekh AD, Ranck MC, Golden DW, Kumar KA, Sood RF, et al. RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation. Proc Natl Acad Sci U S A. 2014;111(4):E484–91.
Wei ZW, Xia GK, Wu Y, Chen W, Xiang Z, Schwarz RE, et al. CXCL1 promotes tumor growth through VEGF pathway activation and is associated with inferior survival in gastric cancer. Cancer Lett. 2015;359(2):335–43.
Nikitovic D, Kouvidi K, Voudouri K, Berdiaki A, Karousou E, Passi A, et al. The motile breast cancer phenotype roles of proteoglycans/glycosaminoglycans. Biomed Res Int. 2014;2014:124321.
Wang Y, Li Z, Zhang H, Jin H, Sun L, Dong H, et al. HIF-1alpha and HIF-2alpha correlate with migration and invasion in gastric cancer. Cancer Biol Ther. 2010;10(4):376–82.
Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–51.
Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–65.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8.
Swanton C, Caldas C. Molecular classification of solid tumours: towards pathway-driven therapeutics. Br J Cancer. 2009;100(10):1517–22.
Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–61.
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74(23):6890–902.
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–26.
Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–54.
Acknowledgments
This study was financially supported by grants from the National Natural Science Foundation of China (Nos. 91529302, 81572798 and 81272749), the Key Projects in the National Science & Technology Pillar Program of China (No. 2014BAI09B03), the Science and Technology Fund of Shanghai Jiao Tong University School of Medicine (No. 13XJ10011) and the Shanghai Jiao Tong University Medical Engineering Cross Research Fund (No. YG2014MS59).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
None
Additional information
Zhi-yuan Fan and Wentao Liu are contributed equally to this work
Electronic supplementary material
Table S1
The 208 common lncRNAs in the list. (XLS 35 kb)
Table S2
The re-annotated lncRNAs listed in the 2 mRNA platforms. (XLSX 218 kb)
Table S3
Weight value of the 35 lncRNAs in the 3 linear classifiers in the training set. (XLSX 14 kb)
Table S4
Detailed results of the reclassification of the samples using different classifiers in the training set. (XLSX 21 kb)
Table S5
Detailed results of the reclassification of the samples using different classifiers in the test data sets. (XLSX 37 kb)
Table S6
Variable and relative importance of the 30 lncRNAs using the random survival forest algorithm. (XLSX 12 kb)
Table S7
Detailed information of differentially expressed genes between the low-risk and high-risk groups. (XLSX 488 kb)
Table S8
GSEA results of each of the 5 lncRNAs. (XLSX 145 kb)
Fig. S1
ROC curves of the 5-lncRNA signature in diagnosing GC in GSE63089. (DOCX 129 kb)
Fig. S2
ROC curves of the 5-lncRNA signature in diagnosing GC in GSE27342. (DOCX 131 kb)
Fig. S3
ROC curves of the 5-lncRNA signature in diagnosing GC in GSE50710. (DOCX 132 kb)
Fig. S4
Melting curves of qRT-PCR for AK001094, AK024171, AK093735, BC003519 and NR_003573 and GAPDH. (a) Melting curves of qRT-PCR for AK001094. (GIF 178 kb)
Figure S4
(b) Melting curves of qRT-PCR for AK024171.(GIF 220 kb)
Figure S4
(c) Melting curves of qRT-PCR for AK093735. (GIF 233 kb)
Figure S4
(d) Melting curves of qRT-PCR for BC003519. (GIF 200 kb)
Figure S4
(e) Melting curves of qRT-PCR for NR_003573. (GIF 204 kb)
ESM 5
(f) Melting curves of qRT-PCR for GAPDH. (GIF 221 kb)
Rights and permissions
About this article
Cite this article
Fan, Zy., Liu, W., Yan, C. et al. Identification of a five-lncRNA signature for the diagnosis and prognosis of gastric cancer. Tumor Biol. 37, 13265–13277 (2016). https://doi.org/10.1007/s13277-016-5185-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5185-9