Abstract
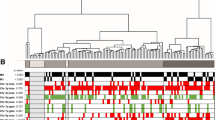
Epigenetic alterations in the methylome have been associated with tumor development and progression in renal cell carcinoma (RCC). In this study, 45 tumor samples, 12 tumor-free kidney cortex tissues, and 24 peripheral blood samples from patients with clear cell RCC (ccRCC) were analyzed by genome-wide promoter-directed methylation arrays and related to clinicopathological parameters. Unsupervised hierarchical clustering separated the tumors into two distinct methylation groups (clusters A and B), where cluster B had higher average methylation and increased number of hypermethylated CpG sites (CpGs). Furthermore, tumors in cluster B had, compared with cluster A, a larger tumor diameter (p = 0.033), a higher morphologic grade (p < 0.001), a higher tumor-node-metastasis (TNM) stage (p < 0.001), and a worse prognosis (p = 0.005). Higher TNM stage was correlated to an increase in average methylation level (p = 0.003) and number of hypermethylated CpGs (p = 0.003), whereas a number of hypomethylated CpGs were mainly unchanged. However, the predicted age of the tumors based on methylation profile did not correlate with TNM stage, morphological grade, or methylation cluster. Differently methylated (DM) genes (n = 840) in ccRCC samples compared with tumor-free kidney cortex samples were predominantly hypermethylated and a high proportion were identified as polycomb target genes. The DM genes were overrepresented by transcription factors, ligands, and receptors, indicating functional alterations of significance for ccRCC progression. To conclude, increased number of hypermethylated genes was associated with increased TNM stage of the tumors. DNA methylation classification of ccRCC tumor samples at diagnosis can serve as a clinically applicable prognostic marker in ccRCC.




Similar content being viewed by others
References
Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev. 2008;34(3):193–205. doi:10.1016/j.ctrv.2007.12.001.
Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–59. doi:10.1056/NEJMra072067.
Banks RE, Tirukonda P, Taylor C, Hornigold N, Astuti D, Cohen D, et al. Genetic and epigenetic analysis of von Hippel-Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer Res. 2006;66(4):2000–11. doi:10.1158/0008-5472.can-05-3074.
Sobin LH GM, Wittekind C. TNM classification of malignant tumors. UICC International Union Against Cancer. New York: Wiley-Blackwell; 2009.
Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6(7):655–63.
Thorstenson A, Bergman M, Scherman-Plogell AH, Hosseinnia S, Ljungberg B, Adolfsson J, et al. Tumour characteristics and surgical treatment of renal cell carcinoma in Sweden 2005–2010: a population-based study from the National Swedish Kidney Cancer Register. Scand J Urol. 2014;48(3):231–8. doi:10.3109/21681805.2013.864698.
Athar U, Gentile TC. Treatment options for metastatic renal cell carcinoma: a review. Can J Urol. 2008;15(2):3954–66.
Morris MR, Maher ER. Epigenetics of renal cell carcinoma: the path towards new diagnostics and therapeutics. Genome Med. 2010;2(9):59. doi:10.1186/gm180.
Ricketts CJ, Hill VK, Linehan WM. Tumor-specific hypermethylation of epigenetic biomarkers, including SFRP1, predicts for poorer survival in patients from the TCGA Kidney Renal Clear Cell Carcinoma (KIRC) project. PLoS One. 2014;9(1):e85621. doi:10.1371/journal.pone.0085621.
Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91(21):9700–4.
Dulaimi E, Ibanez de Caceres I, Uzzo RG, Al-Saleem T, Greenberg RE, Polascik TJ, et al. Promoter hypermethylation profile of kidney cancer. Clin Cancer Res. 2004;10(12 Pt 1):3972–9. doi:10.1158/1078-0432.ccr-04-0175.
Morrissey C, Martinez A, Zatyka M, Agathanggelou A, Honorio S, Astuti D, et al. Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res. 2001;61(19):7277–81.
Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, Khalili H, et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30(12):1390–401. doi:10.1038/onc.2010.525.
Dahl E, Wiesmann F, Woenckhaus M, Stoehr R, Wild PJ, Veeck J, et al. Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene. 2007;26(38):5680–91. doi:10.1038/sj.onc.1210345.
Ricketts CJ, Morris MR, Gentle D, Brown M, Wake N, Woodward ER, et al. Genome-wide CpG island methylation analysis implicates novel genes in the pathogenesis of renal cell carcinoma. Epigenetics. 2012;7(3):278–90. doi:10.4161/epi.7.3.19103.
Morris MR, Ricketts C, Gentle D, Abdulrahman M, Clarke N, Brown M, et al. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29(14):2104–17. doi:10.1038/onc.2009.493.
Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;4:503–18. United States.
Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;2:301–13. United States.
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;10:R115. England.
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–67. doi:10.1016/j.molcel.2012.10.016.
Svenson U, Ljungberg B, Roos G. Telomere length in peripheral blood predicts survival in clear cell renal cell carcinoma. Cancer Res. 2009;69(7):2896–901. doi:10.1158/0008-5472.can-08-3513.
Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–36. doi:10.1093/nar/gki987.
Kohn L, Svenson U, Ljungberg B, Roos G. Specific genomic aberrations predict survival, but low mutation rate in cancer hot spots, in clear cell renal cell carcinoma. Appl Immunohistochem Mol Morphol. 2014. doi:10.1097/pai.0000000000000087.
Ward Jr JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58(301):236–44.
Li J, Jin H, Wang X. Epigenetic biomarkers: potential applications in gastrointestinal cancers. ISRN Gastroenterol. 2014;2014:464015. doi:10.1155/2014/464015.
Lai RK, Chen Y, Guan X, Nousome D, Sharma C, Canoll P, et al. Genome-wide methylation analyses in glioblastoma multiforme. PLoS One. 2014;9(2), e89376. doi:10.1371/journal.pone.0089376.
Barrow TM, Michels KB. Epigenetic epidemiology of cancer. Biochem Biophys Res Commun. 2014. doi:10.1016/j.bbrc.2014.08.002.
Borssen M, Palmqvist L, Karrman K, Abrahamsson J, Behrendtz M, Heldrup J, et al. Promoter DNA methylation pattern identifies prognostic subgroups in childhood T-cell acute lymphoblastic leukemia. PLoS One. 2013;8(6), e65373. doi:10.1371/journal.pone.0065373.
Arai E, Chiku S, Mori T, Gotoh M, Nakagawa T, Fujimoto H, et al. Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis. 2012;33(8):1487–93. doi:10.1093/carcin/bgs177.
Arai E, Kanai Y, Ushijima S, Fujimoto H, Mukai K, Hirohashi S. Regional DNA hypermethylation and DNA methyltransferase (DNMT) 1 protein overexpression in both renal tumors and corresponding nontumorous renal tissues. Int J Cancer. 2006;119(2):288–96. doi:10.1002/ijc.21807.
Halin S, Hammarsten P, Adamo H, Wikstrom P, Bergh A. Tumor indicating normal tissue could be a new source of diagnostic and prognostic markers for prostate cancer. Expert Opin Med Diagn. 2011;5(1):37–47. doi:10.1517/17530059.2011.540009.
Huang H, Tang Y, He W, Huang Q, Zhong J, Yang Z. Key pathways and genes controlling the development and progression of clear cell renal cell carcinoma (ccRCC) based on gene set enrichment analysis. Int Urol Nephrol. 2014;46(3):539–53. doi:10.1007/s11255-013-0511-2.
Maruschke M, Reuter D, Koczan D, Hakenberg OW, Thiesen HJ. Gene expression analysis in clear cell renal cell carcinoma using gene set enrichment analysis for biostatistical management. BJU Int. 2011;108(2 Pt 2):E29–35. doi:10.1111/j.1464-410X.2010.09794.x.
Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–9. doi:10.1038/nature09784.
Liu L, Xu Z, Zhong L, Wang H, Jiang S, Long Q, et al. Prognostic value of EZH2 expression and activity in renal cell carcinoma: a prospective study. PLoS One. 2013;8(11), e81484. doi:10.1371/journal.pone.0081484.
Wang Y, Chen Y, Geng H, Qi C, Liu Y, Yue D. Overexpression of YB1 and EZH2 are associated with cancer metastasis and poor prognosis in renal cell carcinomas. Tumour Biol. 2015;36(9):7159–66. doi:10.1007/s13277-015-3417-z.
Degerman S, Landfors M, Siwicki JK, Revie J, Borssen M, Evelonn E, et al. Immortalization of T-cells is accompanied by gradual changes in CpG methylation resulting in a profile resembling a subset of T-cell leukemias. Neoplasia. 2014;16(7):606–15. doi:10.1016/j.neo.2014.07.001.
Horvath S. Erratum to: DNA methylation age of human tissues and cell types. Genome Biol 2015;16:96.
Wagener N, Holland D, Bulkescher J, Crnkovic-Mertens I, Hoppe-Seyler K, Zentgraf H, et al. The enhancer of zeste homolog 2 gene contributes to cell proliferation and apoptosis resistance in renal cell carcinoma cells. Int J Cancer. 2008;123(7):1545–50. doi:10.1002/ijc.23683.
Wagner W, Weidner CI, Lin Q. Do age-associated DNA methylation changes increase the risk of malignant transformation? Bioessays. 2015;37(1):20–4. doi:10.1002/bies.201400063.
Acknowledgments
This study was supported by grants from the Swedish Cancer Society (BL, GR), the Cancer Research Foundation in Umeå (BL, GR, SD), the Kempe Foundations (GR, SD), and the Västerbotten County Council (BL, GR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Ethical approval
The study was approved by the regional ethical review board in Umeå (Dnr 2011-156-31M 110523).
Informed consent
Informed consent was obtained from all participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Preprocessing of methylation data from Illumina Infinium HumanMeth27K arrays. CpG sites were excluded if ≤ 3 reported beads/array in any sample, if CpG sites had a detection p value equal to or greater than 0.05 and if CpG sites were located on sex chromosomes X and Y. (GIF 15 kb)
Supplementary Figure 2
Distribution of DM-CpGs, genomic aberrations and mutations in the VHL gene. White blocks represent unmethylated, black methylated, light gray wild type and dark gray deleted/mutated. (GIF 53 kb)
Supplementary Table 1
(DOCX 100 kb)
Rights and permissions
About this article
Cite this article
Evelönn, E.A., Degerman, S., Köhn, L. et al. DNA methylation status defines clinicopathological parameters including survival for patients with clear cell renal cell carcinoma (ccRCC). Tumor Biol. 37, 10219–10228 (2016). https://doi.org/10.1007/s13277-016-4893-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-4893-5