Abstract
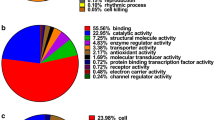
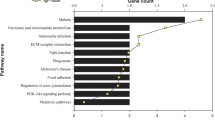
Esophageal squamous cell carcinoma (ESCC) is one of the most common cancers. In this study, our objective was to identify differentially regulated proteins in ESCC using isobaric tag for relative and absolute quantification (iTRAQ) technique and liquid chromatography–tandem mass spectrometry (LC–MS/MS). We compared the protein expression profiles of ESCC tumor tissues with the corresponding adjacent normal tissue from three patients. It was determined that 72 and 57 unique proteins were significantly up-regulated and down-regulated in all three samples. In addition, there were 431 significantly differentially regulated proteins having at least two biological samples. This subject found some of the differential proteins, such as prolyl 4-hydroxylase subunit alpha-1, prolyl 4-hydroxylase subunit alpha-2, and calponin-2, immunoglobulin superfamily containing leucine-rich repeat protein, and prolyl 3-hydroxylase1, which were few studies about them in ESCC. In order to determine the results, we performed another independent experiment. Our results indicated quantitative proteomics, as a robust discovery tool for the identification, differentially regulated proteins in cancers.






Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Zhang P, Xi M, Li Q-Q, He L-R, Liu S-L, Zhao L, et al. The modified Glasgow prognostic score is an independent prognostic factor in patients with inoperable thoracic esophageal squamous cell carcinoma undergoing chemoradiotherapy. J Cancer. 2014;5(8):689–95.
Keszei AP, Alexandra Goldbohm R, Schouten LJ, Jakszyn P, van den Brandt PA. Dietary N-nitroso compounds, endogenous nitrosation, and the risk of esophageal and gastric cancer subtypes in the Netherlands. Am J Clin Nutr. 2013;97(1):135–46.
D’Journo XB, Thomas PA. Current management of esophageal cancer. J Thorac Dis. 2014;6 Suppl 2:S253–64.
Guo X, Hao Y, Kamilijiang M, Hasimu A, Yuan J, Wu G, et al. Potential predictive plasma biomarkers for cervical cancer by 2D-DIGE proteomics and ingenuity pathway analysis. Tumour Biol. 2015;36(3):1711–20.
Atrih A, Mudaliar MA, Zakikhani P, Lamont DJ, Huang JT, Bray SE, et al. Quantitative proteomics in resected renal cancer tissue for biomarker discovery and profiling. Br J Cancer. 2014;110(6):1622–33.
Gan CS, Chong PK, et al. Technical, experimental, and biological variations in isobaric tags for relatives and absolute quantitation (iTRAQ). J Proteome Res. 2007;6(2):821–7.
Gilmore JM, Washburn MP. Advances in shotgun proteomics and the analysis of membrane proteomes. J Proteomics. 2010;73(11):2078–91.
Xiao Z, Li G, Chen Y, Li M, Peng F, et al. Quantitative proteomic analysis of formalin-fixed and paraffin-embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography and tandem mass spectrometry. J Histochem Cytochem. 2010;58(6):517–27.
Finn RD, Marshall M, Bateman A. Pfam: visualization of protein–protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21(3):410–2.
Saccone G, Louis C, Zhang H, Petrella V, Di Natale M, Perri M, et al. Male-specific phosphorylated SR proteins in adult flies of the Mediterranean fruitfly Ceratitis capitata. BMC Genetics. 2014;15 Suppl 2:S6.
Xu Q, Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31(19):5635–43.
Sanford JR, Gray NK, Beckmann K, Cáceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18(7):755–68.
Fortes P, Longman D, McCracken S, Ip JY, Poot R, Mattaj IW, et al. Identification and characterization of RED120: a conserved PWI domain protein with links to splicing and 3′-end formation. FEBS Lett. 2007;581:3087–97.
Zhou A, Ou AC, Cho A, Benz Jr EJ, Huang SC. Novel splicing factor RBM25 modulates Bcl-x Pre-mRNA 5′ splice site selection. Mol Cell Biol. 2008;28(19):5924–36.
Gilkes DM, Saumendra B, Pallavi C, Denis W, Semenza GL. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem. 2013;288(15):10819–29.
Zhang K, Corsa CA, Ponik SM, Prior JL, et al. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol. 2013;15(6):677–87.
Gilkes DM, Chaturvedi P. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73(11):3285–96.
Chakravarthi BV, Pathi SS, Goswami MT, et al. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5(16):6654–69.
Pan PW, Zhang Q, Bai F, Hou J, Bai G. Profiling and comparative analysis of glycoproteins in Hs578BST and Hs578T and investigation of prolyl 4-hydroxylase alpha polypeptide II expression and influence in breast cancer cells. Biochemistry (Mosc). 2012;77(5):539–45.
Jarzab B, Wiench M, Fujarewicz K, Simek K, et al. Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Res. 2005;65(4):1587–97.
Chang KP, Yu JS, Chien KY, Lee CW, Liang Y, et al. Identification of PRDX4 and P4HA2 as metastasis associated proteins in oral cavity squamous cell carcinoma by comparative tissue proteomics of microdissected specimens using iTRAQ technology. J Proteome Res. 2011;10(11):4935–47.
Ridley AJ. Rho GTPases and actin dynamics in membrane pro-trusions and vesicle trafficking. Trends Cell Biol. 2006;16(10):522–9.
Kaneko M, Takeoka M, Oguchi M, Koganehira Y, et al. Calpon in hl suppresses tumor growth of src-induce d transformed 3Y1 cells in association with a de crease in angiogenesis. Jpn J Cancer Res. 2002;93(8):935–43.
Yanagisawa Y, Takeoka M, Ehara T, Itano N, Miyagawa S, Taniguchi S. Reduction of calponin H1 expression in human colon cancer blood vessels. Eur J Surg Oncol. 2008;34(5):531–7.
Ogura T, Kobayashi H, Ueoka Y, Okugawa K, Kato K, Hirakawa T, et al. Adenovirus-mediated calponin h1 gene therapy directed against peritoneal dissemination of ovarian cancer: bifunctional therapeutic effects on peritoneal cell layer and cancer cells. Clin Cancer Res. 2006;12(17):5216–23.
Hossain MM, Hwang D-Y, Huang Q-Q, Sasaki Y, Jin J-P. Developmentally regulated expression of calponin isoforms and the effect of h2-calponin on cell proliferation. Am J Physiol. 2003;284(1):156–67.
Choi SY, Jang JH, Kim KR. Analysis of differentially expressed genes in human rectal carcinoma using suppression subtractive hybridization. Clin Exp Med. 2011;11(4):219–26.
Hossain MM, Hwang D-Y, Huang Q-Q, Sasaki Y, Jin J-P. Developmentally regulated expression of calponin isoforms and the effect of h2-calponin on cell proliferation. Am J Physiol. 2003;284(1):C156–67.
Nagasawa A, Kubota R, Imamura Y, Nagamine K, et al. Cloning of the cDNA for a new member of the immunoglobulin superfamily (ISLR) containing leucine-rich repeat (LRR). Genomics. 1997;44(3):273–9.
D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28(12):655–62.
Ishikawa Y, Wirz J, Vranka JA, Nagata K, Bächinger HP. Biochemical characterization of the prolyl 3-hydroxylase 1.cartilage-associated protein.cyclophilin B complex. J Biol Chem. 2009;284(26):17641–7.
Ahmad Mir S, Pavithra R, Jain AP, Khan AA, Datta KK, Mohan SV, et al. LC–MS-based serum metabolomic analysis reveals dysregulation of phosphatidylcholines in esophageal squamous cell carcinoma. J Proteomics. 2015;S1874-3919(15):00231–6.
Li C, Guo X, Jianqing Z, Yang M, Ge B, Li Z. Serum differential protein identification of Xinjiang Kazakh esophageal cancer patients based on the two-dimensional liquid-phase chromatography and LTQ MS. Mol Biol Rep. 2014;41(5):2893–905.
Conflicts of interest
None
Funding
This study was supported by the grant from the National Natural Science Foundation of China, NO: 81260308, and the Open Subject about Xinjiang Major Diseases of China, NO: SKLIB-XJMDR-2014-12.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Feiyan Deng and Keming Zhou contributed equally to this work.
Rights and permissions
About this article
Cite this article
Deng, F., Zhou, K., Li, Q. et al. iTRAQ-based quantitative proteomic analysis of esophageal squamous cell carcinoma. Tumor Biol. 37, 1909–1918 (2016). https://doi.org/10.1007/s13277-015-3840-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3840-1