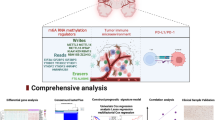
Abstract
Background
Aberrant DNA methylation is one of the major epigenetic alterations in neuroblastoma.
Objective
Exploring the prognostic significance of methylation driver genes in neuroblastoma could help to comprehensively assess patient prognosis.
Methods
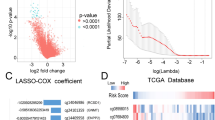
After identifying methylation driver genes (MDGs), we used the LASSO algorithm and stepwise Cox regression to construct methylation driver gene-related risk score (MDGRS), and evaluated its predictive performance by multiple methods. By combining risk grouping and MDGRS grouping, we developed a new prognostic stratification strategy and explored the intrinsic differences between the different groupings.
Results
We identified 44 stably expressed MDGs in neuroblastoma. MDGRS showed superior predictive performance in both internal and external cohorts and was strongly correlated with immune-related scores. MDGRS can be an independent prognostic factor for neuroblastoma, and we constructed the nomogram to facilitate clinical application. Based on the new prognostic stratification strategy, we divided the patients into three groups and found significant differences in overall prognosis, clinical characteristics, and immune infiltration between the different subgroups.
Conclusion
MDGRS was an accurate and promising tool to facilitate comprehensive pre-treatment assessment. And the new prognostic stratification strategy could be helpful for clinical decision making.







Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Data Availability
Publicly available datasets were used in this study. GSE73515 (PRJNA297203), GSE73517 (PRJNA297205), GSE49710 (PRJNA214798) and GSE62564 (PRJNA264621) are available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). E-MTAB-8248 is downloaded from the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/).
References
Aravindan N, Herman T, Aravindan S (2020) Emerging therapeutic targets for neuroblastoma. Expert Opin Ther Targets 24:899–914. https://doi.org/10.1080/14728222.2020.1790528
Bai Y, Wei C, Zhong Y, Zhang Y, Long J, Huang S, Xie F, Tian Y, Wang X, Zhao H (2020) Development and validation of a Prognostic Nomogram for Gastric Cancer based on DNA methylation-driven differentially expressed genes. Int J Biol Sci 16:1153–1165. https://doi.org/10.7150/ijbs.41587
Blanche P, Dartigues JF, Jacqmin-Gadda H (2013) Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med 32:5381–5397. https://doi.org/10.1002/sim.5958
Campbell K, Naranjo A, Hibbitts E, Gastier-Foster JM, Bagatell R, Irwin MS, Shimada H, Hogarty M, Park JR, DuBois SG (2020) Association of heterogeneous MYCN amplification with clinical features, biological characteristics and outcomes in neuroblastoma: a report from the Children’s Oncology Group. Eur J Cancer 133:112–119. https://doi.org/10.1016/j.ejca.2020.04.007
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z (2017) Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18:248–262. https://doi.org/10.1016/j.celrep.2016.12.019
Chen DP, Lin YC, Fann CS (2016) Methods for identifying differentially methylated regions for sequence- and array-based data. Brief Funct Genomics 15:485–490. https://doi.org/10.1093/bfgp/elw018
Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D et al (2009) The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 27:289–297. https://doi.org/10.1200/jco.2008.16.6785
Darvin P, Toor SM, Sasidharan Nair V, Elkord E (2018) Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 50:1–11. https://doi.org/10.1038/s12276-018-0191-1
Decock A, Ongenaert M, Hoebeeck J, De Preter K, Van Peer G, Van Criekinge W, Ladenstein R, Schulte JH, Noguera R, Stallings RL et al (2012) Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biol 13:R95. https://doi.org/10.1186/gb-2012-13-10-r95
Esposito MR, Aveic S, Seydel A, Tonini GP (2017) Neuroblastoma treatment in the post-genomic era. J Biomed Sci 24:14. https://doi.org/10.1186/s12929-017-0319-y
Fetahu IS, Taschner-Mandl S (2021) Neuroblastoma and the epigenome. Cancer Metastasis Rev 40:173–189. https://doi.org/10.1007/s10555-020-09946-y
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized Linear models via Coordinate Descent. J Stat Softw 33:1–22
Guo Y, Xu F, Lu T, Duan Z, Zhang Z (2012) Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev 38:904–910. https://doi.org/10.1016/j.ctrv.2012.04.007
Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14:7. https://doi.org/10.1186/1471-2105-14-7
Henrich KO, Bender S, Saadati M, Dreidax D, Gartlgruber M, Shao C, Herrmann C, Wiesenfarth M, Parzonka M, Wehrmann L et al (2016) Integrative Genome-Scale Analysis Identifies Epigenetic Mechanisms of Transcriptional Deregulation in unfavorable neuroblastomas. Cancer Res 76:5523–5537. https://doi.org/10.1158/0008-5472.Can-15-2507
Koch A, Joosten SC, Feng Z, de Ruijter TC, Draht MX, Melotte V, Smits KM, Veeck J, Herman JG, Van Neste L et al (2018) Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol 15:459–466. https://doi.org/10.1038/s41571-018-0004-4
Lalchungnunga H, Hao W, Maris JM, Asgharzadeh S, Henrich KO, Westermann F, Tweddle DA, Schwalbe EC, Strathdee G (2022) Genome wide DNA methylation analysis identifies novel molecular subgroups and predicts survival in neuroblastoma. Br J Cancer 127:2006–2015. https://doi.org/10.1038/s41416-022-01988-z
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. https://doi.org/10.1186/1471-2105-9-559
Lau DT, Hesson LB, Norris MD, Marshall GM, Haber M, Ashton LJ (2012) Prognostic significance of promoter DNA methylation in patients with childhood neuroblastoma. Clin Cancer Res 18:5690–5700. https://doi.org/10.1158/1078-0432.Ccr-12-0294
Lundberg KI, Treis D, Johnsen JI (2022) Neuroblastoma heterogeneity, plasticity, and emerging therapies. Curr Oncol Rep 24:1053–1062. https://doi.org/10.1007/s11912-022-01270-8
Macheret M, Halazonetis TD (2015) DNA replication stress as a hallmark of cancer. Annu Rev Pathol 10:425–448. https://doi.org/10.1146/annurev-pathol-012414-040424
Maris JM (2010) Recent advances in neuroblastoma. N Engl J Med 362:2202–2211. https://doi.org/10.1056/NEJMra0804577
Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, Pessoa C (2019) Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics 14:1164–1176. https://doi.org/10.1080/15592294.2019.1640546
Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Krämer A, Roncaioli JL, Sand F, Heuckmann JM et al (2015) Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526:700–704. https://doi.org/10.1038/nature14980
Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR et al (2015) Advances in risk classification and treatment strategies for Neuroblastoma. J Clin Oncol 33:3008–3017. https://doi.org/10.1200/jco.2014.59.4648
Skvortsova K, Stirzaker C, Taberlay P (2019) The DNA methylation landscape in cancer. Essays Biochem 63:797–811. https://doi.org/10.1042/ebc20190037
Swift CC, Eklund MJ, Kraveka JM, Alazraki AL (2018) Updates in diagnosis, management, and Treatment of Neuroblastoma. Radiographics 38:566–580. https://doi.org/10.1148/rg.2018170132
Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, Teschendorff AE (2017) ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 33:3982–3984. https://doi.org/10.1093/bioinformatics/btx513
Tolbert VP, Matthay KK (2018) Neuroblastoma: clinical and biological approach to risk stratification and treatment. Cell Tissue Res 372:195–209. https://doi.org/10.1007/s00441-018-2821-2
Wang R, Wang Q (2021) Identification and External Validation of a Transcription Factor-Related Prognostic Signature in Pediatric Neuroblastoma. J Oncol 2021:1370451. https://doi.org/10.1155/2021/1370451
Watanabe K, Kimura S, Seki M, Isobe T, Kubota Y, Sekiguchi M, Sato-Otsubo A, Hiwatari M, Kato M, Oka A et al (2022) Identification of the ultrahigh-risk subgroup in neuroblastoma cases through DNA methylation analysis and its treatment exploiting cancer metabolism. Oncogene 41:4994–5007. https://doi.org/10.1038/s41388-022-02489-2
Whiteside TL (2006) Immune suppression in cancer: effects on immune cells, mechanisms and future therapeutic intervention. Semin Cancer Biol 16:3–15. https://doi.org/10.1016/j.semcancer.2005.07.008
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L et al (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov (Camb) 2:100141. https://doi.org/10.1016/j.xinn.2021.100141
Yang J, Zhou J, Li C, Wang S (2021) Integrated analysis of the functions and prognostic values of RNA-binding proteins in neuroblastoma. PLoS ONE 16:e0260876. https://doi.org/10.1371/journal.pone.0260876
Zafar A, Wang W, Liu G, Wang X, Xian W, McKeon F, Foster J, Zhou J, Zhang R (2021) Molecular targeting therapies for neuroblastoma: Progress and challenges. Med Res Rev 41:961–1021. https://doi.org/10.1002/med.21750
Zhang P, Ma K, Ke X, Liu L, Li Y, Liu Y, Wang Y (2021) Development and validation of a Five-RNA-Based signature and identification of candidate Drugs for Neuroblastoma. Front Genet 12:685646. https://doi.org/10.3389/fgene.2021.685646
Zhang C, Ding Z, Luo H (2022) The Prognostic Role of m6A-Related genes in paediatric neuroblastoma patients. Comput Math Methods Med 2022:8354932. https://doi.org/10.1155/2022/8354932
Funding
This study was supported by grants from the National Natural Science Foundation of China (No: 81502187); Key scientific research projects of colleges and universities in Henan Province (No. 20A320020, Jiao Zhang); the Basic and Frontier Technology Research Project of Henan Province (No. 212102310032, Jiao Zhang).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Jiao Zhang provided ideas and carried out research and design. Yahui Han and Biyun Li completed the material preparation, data collection and data analysis and contributed equally to this paper. The first draft of the manuscript was completed by Jian Cheng and Diming Zhou, modified by Xiafei Yuan, Wei Zhao and Da Zhang, and determined by Jiao Zhang. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
All authors consent to the publication of this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.



Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, Y., Li, B., Cheng, J. et al. Construction of methylation driver gene-related prognostic signature and development of a new prognostic stratification strategy in neuroblastoma. Genes Genom 46, 171–185 (2024). https://doi.org/10.1007/s13258-023-01483-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-023-01483-6