Abstract
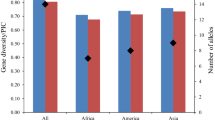
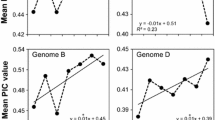
The cultivated soybean [Glycine max (L.) Merr.] is widely considered to descend from the wild soybean (G. soja Sieb. & Zucc.). This study was designed to evaluate the genetic variability and differentiation between G. soja and G. max, and to detect signatures of the selection that may have occurred during the domestication process from G. soja to G. max. A total of 192 G. soja accessions and 104 G. max accessions were genotyped using eight selected simple sequence repeat (SSR) markers assigned to three SSR groups. Four SSRs in group A were not located near any known QTL. Three SSRs in group B were associated with seed protein content, and an SSR in group C was associated with resistance to Sclerotinia stem rot. The number of alleles per locus and the level of genetic variability in G. soja were higher than those in G. max. A total of 122 out of 125 alleles were present in G. soja, but only 59 alleles were detected in G. max. The average gene diversity was 0.74 in G. soja and 0.64 in G. max. Four SSRs near QTLs of agronomic importance showed strong genetic differentiation and shift change in high frequency alleles in groups B and C between G. soja and G. max, revealing selection signatures that may reflect the domestication events and recent selective breeding. With reduced diversity in G. max, some undomesticated genes from G. soja should be prime candidates for introgression to increase the pool of diversity in G. max.
Similar content being viewed by others
References
Arahana VS, Graef GL, Specht JE, Steadman JR and Eskridge KM (2001) Identification of QTLs for Resistance to Sclerotinia sclerotiorum in soybean. Crop Sci. 41: 180–188.
Beja-Pereira A, Luikart G, England PR, Bradley DG, Jann OC, Bertorelle G, Chamberlain AT, Nunes TP, Metodiev S, Ferrand N and Erhardt G (2003) Gene-culture coevolution between cattle milk protein genes and human lactase genes. Nat. Genet. 35: 311–313.
Brinkman MA and Frey KJ (1977) Yield component analysis of oat isolines that produce different grain yield. Crop Sci. 17: 165–168.
Brummer EC, Graef GL, Orf J, Wilcox JR and Shoemaker RC (1997) Mapping QTL for seed protein and oil content in eight soybean populations. Crop Sci. 37: 370–378.
Chen Y and Nelson RL (2004) Genetic variation and relationships among cultivated, wild, and semiwild soybean. Crop Sci. 44: 316–325.
Chen Y and Nelson RL (2005) Relationship between origin and genetic diversity in Chinese soybean germplasm. Crop Sci. 45: 1645–1652.
Chung J, Babka HL, Graef GL, Staswick PE, Lee DJ, Cregan PB, Shoemaker RC and Specht JE (2003) The seed protein, oil, and yield QTL on soybean linkage group I. Crop Sci. 43: 1053–1067.
Concibido VC, La Vallee B, Mclaird P, Pineda N, Meyer J, Hummel L, Yang J, Wu K and Delannay X (2003) Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor. Appl. Genet. 106: 575–582.
Diers BW, Keim P, Fehr WR and Shoemaker RC (1992) RFLP analysis of soybean seed protein and oil content. Theor. Appl. Genet. 83: 608–612.
Excoffier L, Smouse PE and Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 136: 343–359.
Gregory TR (2009) Artificial selection and domestication: Modern lessons from Darwin’s enduring analogy. Evol. Edu. Outreach 2: 5–27.
Hahn V (2002) Genetic variation for resistance to Sclerotinia head rot in sunflower inbred lines. Field Crops Res. 77: 153–159.
Hyten DL, Song Q, Zhu Y, Choi I-Y, Nelson RL, Costa JM, Specht JE, Shoemaker RC and Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 103: 16666–16671.
Jun T-H, Van K, Kim MY, Lee S-H and Walker DR (2008) Association analysis using SSR markers to find QTL for seed protein content in soybean. Euphytica 162: 179–191.
Kim HS and Diers BW (2000) Inheritance of partial resistance to Sclerotinia stem rot in soybean. Crop Sci. 40: 55–61.
Kuroda Y, Kaga A, Tomooka N and Vaughan DA (2006) Population genetic structure of Japanese wild soybean (Glycin soja) based on microsatellite variation. Mol. Ecol. 15: 959–974.
Li Z and Nelson RL (2002) RAPD marker diversity among cultivated and wild soybean accessions from four Chinese provinces. Crop Sci. 42: 1737–1744.
Maynard SJ and Haigh J (1974) The hitchhiking effect of a favorable gene. Genet. Res. 23: 23–35.
Ohara M and Shimamoto Y (2002) Importance of genetic characterization and conservation of plant genetic resources: The breeding system and genetic diversity of wild soybean (Glycine soja). Plant Species Biol. 17: 51–58.
Palaisa KA, Morgante M, Williams M and Rafalski A (2003) Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15: 1795–1806.
Peakall R and Smouse PE (2006) GenAIEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6: 288–295.
Remington DL, Thronsbery JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM and Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484.
Sebolt AM, Shoemaker RC and Diers BW (2000) Analysis of quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Sci. 40: 1438–1444.
Tanksley SD and McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066.
Vigouroux Y, McMullen M, Hittinger CT, Houchins K, Schulz L, Kresovich S, Matsuoka Y and Doebley J (2002) Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc. Natl. Acad. Sci. USA 23: 9650–9655.
Wang D, Arelli PR, Shoemaker RC and Diers BW (2001) Loci underlying resistance to Race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor. Appl. Genet. 103: 561–566.
Wang D, Graef GL, Procopiuk AM and Diers BW (2004) Identification of putative QTL that underlie yield in interspecific soybean backcross populations. Theor. Appl. Genet. 108: 458–467.
Wang RL, Stec A, Hey J, Lukens L and Doebley J (1999) The limits of selection during maize domestication. Nature 398: 236–239.
Winter SMJ, Shelp BJ, Anderson TR, Welacky TW and Rajcan I (2007) QTL associated with horizontal resistance to soybean cyst nematode in Glycine soja PI464925B. Theor. Appl. Genet. 114: 461–472.
Xu DH, Abe J, Gai JY and Shimamoto Y (2002) Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theor. Appl. Genet. 105: 645–653.
Xu DH and Gai JY (2003) Genetic diversity of wild cultivated soybeans growing in China revealed by RAPD analysis. Plant Breed. 122: 503–506.
Zeder MA, Emshwiller E, Smith BD and Bradley DG (2006) Documenting domestication: the intersection of genetics and archaeology. Trend. Genet. 22: 139–155.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Jun, TH., Van, K., Kim, M.Y. et al. Uncovering signatures of selection in the soybean genome using SSR diversity near QTLs of agronomic importance. Genes Genom 33, 391–397 (2011). https://doi.org/10.1007/s13258-010-0159-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-010-0159-6