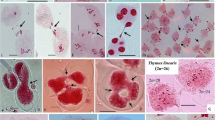
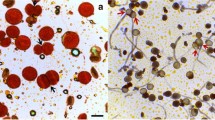
Abstract
Polyploidy is an important event and major force in plant speciation. Amongst the polyploids, allopolyploids have attracted special attention to investigate genetic and epigenetic mechanisms. Also, they are the means for the development of new genotypes and genomic combinations to facilitate genetic enhancement and agricultural productivity. Whereas natural allopolyploids are genetically stable and well adapted, the newly synthesized ones are highly unstable. This instability is manifested into alterations at genomic and/or phenotypic level. Here we present the phenomenon of direct chromosome/chromatin elimination from pollen mother cells (PMCs) in wheat-rye hybrids as one aspect of instability leading to irregular meiosis and disturbances in meiotic process. One of the prominent irregularities noticed is peripherally separated uncondensed or pycnotic masses of chromatin in all meiotic stages. We have observed that this chromatin undergoes elimination by budding-like way, whereby a “mini-cell” is created. It was also found that nucleoli are the first to be eliminated along with a small mass of chromatin. By means of GISH we have shown that both rye and wheat chromatin might be eliminated. In the separated groups of chromosomes/chromatin neither DNaseI nor DNase II activity was detected. Immunolocalization of tubulin allowed for differentiation between chromatin elimination from microspores and elimination from earlier stages of meiosis. It was noticeable, that in microspores special cytoskeleton structure pushing micronuclei out from the cells was created. Elimination occurred before and after meiosis as well as in each stage of meiotic division, but its intensity varied, depending on the PMC. The basis of the elimination mechanism might be the same as in cytomixis, because both phenomena share common symptoms, although cytomixis per se was rare in the analyzed hybrids.





Similar content being viewed by others
References
Baptista-Giacomelli FR, Pagliarini MS, De Almeid JL. Elimination of micronuclei from microspores in Brazilian oat (Avena sativa L.) variety. Genet Mol Biol. 2000;23:681–4.
Barclay IR. High-frequencies of haploid production in wheat (Triticum aestivum) by chromosome elimination. Nature. 1975;256:410–1.
Bennett MD, Finch RA, Barclay IR. The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma. 1976;54:175–200.
Bennett MD, Smith JS. Nuclear DNA amounts in angiosperms. Philos T Roy Soc B. 1976;274:227–74.
Bento M, Pereira HS, Rocheta M, Gustafson P, Viegas W, Silva M. Polyploidization as a retraction force in plant genome evolution: sequence rearrangements in triticale. PLoS Genet. 2008;3(1):e1402. doi:10.1371/journal.pone.0001402.
Bhat TA, Parrveen S, Khan AH. MMS-induced cytomixis in pollen mother cells of broad bean (Vicia faba L.). Turk J Bot. 2006;30:273–9.
Bourc’his D, Bestor T. Meiotic catastrophe and retrotransposons reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9.
Boyko EV, Badaev NS, Maximov NG, Zelenin AV. Does DNA content change in the course of triticale breeding? Cereal Res Commun. 1984;12:99–100.
Brasileiro-Vidal AC, Cuadrado A, Brammer SP, Benko-Iseppon AM, Guerra M. Molecular cytogenetic characterization of parental genomes in the partial amphiploid Triticum aestivum x Thinopyrum ponticum. Gen Mol Biol. 2005;28(2):308–13.
Chen FQ, Hayes PM. Wide hybridization of Hordeum vulgare x Zea mays. Genome. 1991;34:603–5.
Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406.
Chen ZJ, Comai L, Pikaard CS. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc Natl Acad Sci USA. 1998;95:14891–6.
Chen ZJ, Ni Z. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays. 2006;28:240–52.
Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Gene Dev. 1997;11:2124–36.
Chen ZJ, Pikaard CS. Transcriptional analysis of nucleolar dominance in polyploidy plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci USA. 1997;94:3443–7.
Comai L. Genetic and epigenetic interactions in allopolyploid plants. Plant Mol Biol. 2000;43:387–99.
Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, et al. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell. 2000;12:1551–68.
De M, Sharma AK. Cytomixis in pollen mother cells of an apomictic ornamental Ervalamia divaricala (Linn.) Alston. Cytologia. 1983;48:201–7.
De Nettancourt D, Grant WF. La Cytogénétique de Lotus (Leguminosae) III. Un cas de cytomixie dans un hybride interspésifique. Cytologia. 1964;29:191–5.
Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM. Rapid elimination of low-copy DNA sequences in polyploidy wheat: A possible mechanism for differentiation of homoeologous chromosome. Genetics. 1997;147:1381–7.
Finch RA. Tissue-specific elimination of alternative whole parental genomes in one barley hybrid. Chromosoma. 1983;88:386–93.
Flavell RB. The structure and control of expression of ribosomal RNA genes. Plant Mol Cell Biol. 1986;3:252–74.
Furuka Y, Nishikawa K, Tanino T. Stability in DNA content of AB genome component of common wheat during the past seven thousand years. J Genet. 1974;49(4):179–87.
Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Brüß C, et al. Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell. 2005;17:2431–8.
Ghanima AM, Talaat AA. Cytomixis and its possible evolutionary role in Kuwait population of Diplotaxis harra (Boraginaceae). Bot J Linn Soc. 2003;143:169–75.
Goday C, Ruiz MF. Differential acethylation of histones H3 and H4 in paternal and maternal germline chromosomes during development of sciarid flies. J Cell Sci. 2002;115:4765–75.
Gottschalk W. Chromosome and nucleus migration during microsporogenesis of Pisum sativum. Nucleus. 1970;13:1–9.
Gupta SB. Duration of mitotic cycle and regulation of DNA replication in Nicotiana plumbaginifolia and a hybrid derivative of N. tabacum showing chromosome instability. Can J Genet Cytol. 1969;11:133–42.
Hammatt N, Blackall NW, Davey MR. Variation in the DNA content of Glycine species. J Exp Bot. 1991;42:659–65.
Hegarty MJ, Jones JM, Wilson ID, Barker GL, Coghill JA, Sanchez-Baracaldo P, et al. Development of anonymous cDNA microarrays to study changes to the Sencio floral transcriptome during hybrid speciation. Mol Ecol. 2005;14:2493–510.
Jin WW, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, et al. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell. 2004;16:571–81.
Kalinka A, Achrem M, Rogalska S. Application of BSP method in methylation pattern comparison of reverse transcriptase (rt) gene in wheat-rye hybrids and their parental species. In: Naganowska B, Kachlicki P, Krajewski P, editors. Genetyka i genomika w doskonaleniu roślin uprawnych. Institute of Plant Genetics Poznań; 2009. p. 53–61.
Kasha KJ, Kao KN. High frequency haploid production in barley (Hordeum vulgare L). Nature. 1970;225:874–5.
Kloc M, Zagrodzinska B. Chromatin elimination—an oddity or a common mechanism in differentiation and development? Differentiation. 2001;68:84–91.
Komarova NY, Grabe T, Huigen DJ, Hemleben V, Volkov RA. Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Mol Biol. 2004;56:439–63.
Laurie DA, Bennett MD. The timing of chromosome elimination in hexaploid wheat x maize crosses. Genome. 1988;32:953–61.
Laurie DA, Bennett MD. Wheat x maize hybridization. Can J Genet Cytol. 1986;28:313–6.
Linde-Laursen I, von Bothmer R. Orderly arrangement of the chromosomes within barley genomes of chromosome-eliminating Hordeum lechleri x barley hybrids. Genome. 1999;42:225–36.
Liu B, Brubaker CL, Mergeai G, Cronn RC, Wendel JF. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome. 2001;44:321–30.
Liu B, Vega JM, Segal G, Abbo S, Rodova M, Feldman M. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops I Changes in low-copy noncoding DNA sequences. Genome. 1998;41:272–7.
Liu JH, Xu XY, Deng XX. Intergeneric somatic hybridization and its application to crop genetic improvement. Plant Cell Tiss Org. 2005;82:19–44.
Lukens LN, Pires JC, Leon E, Vogelzang R, Oslach L, Osborn T. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 2006;140(1):336–48.
Ma XF, Gustafson JP. Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenet Genome Res. 2005;109:236–49.
Ma XF, Gustafson P. Allopolyploidization-accomodated genomic sequence changes in triticale. Ann Bot. 2008;101:825–32.
Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 2002;129:733–46.
Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, Doerge RW, et al. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005;41:221–30.
Marfil CF, Masuelli RW, Davison J, Comai L. Genomic instability in Solanum tuberosum x Solanum kurtzianum interspecific hybrids. Genome. 2006;49:104–13.
Masterson J. Stomatal size in fossil plants. Evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–4.
Matzk F. Hybrids crosses between oat and Andropogene or Paniceae species. Crop Sci. 1996;36:17–21.
Matzk F, Mahn A. Improved techniques for haploid production in wheat using chromosome elimination. Plant Breed. 1994;113:125–9.
Matzk F, Oertel C, Altenhofer P, Schubert I. Manipulation of reproductive systems in Poaceae to increase the efficiency in crop breeding and production. Trends Agron. 1997;1:19–34.
McClintock B. The relationship of a particular chromosomal element to the development of the nucleoli in Zea mays. Z Zellforsch Mik Ana. 1934;21:294–328.
Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–8.
Morikawa T, Leggett M. Cytological and morphological variations in wild populations of Avena canariensis from Canary Islands. Genes Genet Syst. 1996;71:15–21.
Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–6.
Narayan RKJ. The role of genomic constraints upon evolutionary changes in genome size and chromosome organization. Ann Bot. 1998;82:57–66.
Natali L, Giordani T, Polizzi E, Pugliesi C, Fambrini M, Cavallini A. Genomic alterations in the interspecific hybryd Helianthus annuus x Helianthus tuberosus. Theor Appl Genet. 1998;97:1240–7.
Navashin M. Chromosomal alterations caused by hybridization and their bearing upon certain genetic problems. Cytologia. 1934;6:169–203.
Ohri D, Fritsch RM, Hanelt P. Evolution of genome size in Allium (Alliaceae). Plant Syst Evol. 1998;210:57–86.
Ozkan H, Levy AA, Feldman M. Allopolyploidy-induced rapid genome evolution in wheat (Aegilops-Triticum) group. Plant Cell. 2001;13:1735–47.
Ozkan H, Tuna M, Arumuganathan K. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. J Hered. 2003;94(3):260–4.
Pickering RA. Partial control of chromosome elimination by temperature in immature embryos of Hordeum vulgare L. x H. bulbosum. Euphytica. 1985;14:869–74.
Pikaard CS. Nucleolar dominance and silencing of transcription. Trends Plant Sci. 1999;4:478–83.
Price HJ, Chambers KL, Bachmann K, Riggs J. Inheritance of nuclear 2C DNA content variation in intraspecific and interspecific hybrids of Microseris (Asteraceae). Am J Bot. 1985;70:1133–8.
Reeder RH. Mechanisms of nucleolar dominance in animals and plants. J Cell Biol. 1985;101:2013–6.
Riera-Lizarazu O, Rines HW, Phillips RL. Cytological and molecular characterization of oat x maize partial hybrids. Theor Appl Genet. 1996;93:123–35.
Rines HW, Dahleen LS. Haploids of plants produced by application of maize pollen to emasculated oat florets. Crop Sci. 1990;30:1073–8.
Schranz ME, Osborn TC. Novel flowering time variation in the resynthesized polyploidy Brassica napus. J Hered. 2000;91:242–6.
Schwarzacher-Robinson T, Finch RA, Smith JB, Bennett MD. Genotypic control of centromere positions of parental genomes in Hordeum x Secale hybrid metaphases. J Cell Sci. 1987;87:291–304.
Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell. 2001;13:1749–59.
Shimizu N, Itoh N, Utiyama H, Wahl GM. Selective entrapment of extrachromosomally amplified DNA by nuclear budding and micronucleation during S phase. J Cell Biol. 1998;140:1307–20.
Singhal VK, Gill B. Cytomixis in some woody species. Biologica. 1985;1:168–75.
Singhal VK, Gill B, Dhaliwal RS. Status of chromosomal diversity in the hardwood tree species of Punjab state. J Cytol Genet. 2007;8:67–83.
Singhal VK, Kumar P. Impact of cytomixis on meiosis, pollen viability and pollen size in wild populations of Himalayan poppy (Meconopsis aculeata Royle). J Biosci. 2008;33(3):371–80.
Soltis DE, Soltis PS. Polyploidy: origins of species and genome evolution. Trends Ecol Evol. 1999;9:348–52.
Subrahmanyam NC. Haploidy from Hordeum interspecific crosses I Polyhaploids of H. parodii and H. procerum. Theor Appl Genet. 1977;49:209–17.
Tanaka T, Shimizu N. Induced detachment of acentric chromatin from mitotic chromosomes leads to their cytoplasmic localization at G(1) and the micronucleation by lamin reorganization at S phase. J Cell Sci. 2000;113:697–707.
Taverna SD, Coyne RS, Allis CD. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell. 2002;110:701–11.
Verma SC, Rees H. Nuclear DNA and evolution of allotetraploid Brassicaceae. Heredity. 1974;33:61–8.
Wallace H, Landgridge WHR. Differential amphiplasty and the control of ribosomal RNA synthesis. Heredity. 1971;27:1–13.
Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, et al. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–73.
Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–49.
Wheatley WG, Kasha KJ. Chromosome elimination in bi-nucleate cells of a (2X) H. vulgare X (2X0 H. bulbosum hybrid Barley. Genet Newsl. 1982;12:74–7.
Wolfe KH. Yesterday’s polyploids and the mystery of polyploidization. Nat Rev Genet. 2001;2:333–41.
Zenkteler M, Nitzsche W. Wide hybridization experiments in cereals. Theor Appl Genet. 1984;68:311–5.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kalinka, A., Achrem, M. & Rogalska, S.M. Cytomixis-like chromosomes/chromatin elimination from pollen mother cells (PMCs) in wheat-rye allopolyploids. Nucleus 53, 69–83 (2010). https://doi.org/10.1007/s13237-010-0002-0
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-010-0002-0