Abstract
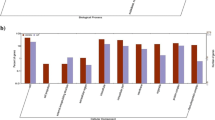
The genus Listeria includes eight different species, but only Listeria monocytogenes is associated to human illness. This microorganism is capable of growing in temperatures ranging from 4 °C to 37 °C and forming biofilms on processing sites, which is of great concern in the food industry. In this current work, the transcription of genes related to biofilm formation, stress-response, and virulence in two strains of L. monocytogenes, serotypes 1/2a and 4b, growing at 7 °C and 37 °C, was analyzed by quantitative real time PCR (qPCR). For both serotypes, a temperature of 7 °C significantly increased (P < 0.05) the transcription level of sigB, prfA, luxS, sufS, sufU, ltrC and flaA genes when compared to growth at 37 °C, whereas transcription of the hly gene did not vary significantly at the temperatures tested. Incubation at 7 °C increased the transcriptional level of the actA gene only in L. monocytogenes serotype 1/2a. On the other hand, L. monocytogenes serotype 4b showed significantly increased transcription of the degU gene at 7 °C. Interestingly, expression of the agrA gene, which is involved in adhesion and biofilm formation on abiotic surfaces, was not detected in serotype 4b, and its transcription level was lower at 7 °C in serotype 1/2a. These results demonstrate that the two studied L. monocytogenes serotypes, which are responsible for many human listeriosis cases, have different molecular mechanisms at temperatures of 7 °C and 37 °C.

Similar content being viewed by others
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Becker LA, Evans SN, Hutkins RW, Benson AK (2000) Role of sigma(B) in adaption of Listeria monocytogenes to growth at low temperature. J Bacteriol 182:7083–7087
Ben Slama R, Miladi H, Chaieb K, Bakhrouf A (2013) Survival of Listeria monocytogenes cells and the effect of extended frozen storage (−20 °C) on the expression of its virulence gene. Appl Biochem Biotechnol 170:1174–1183. doi:10.1007/s12010-013-0253-8
Borucki MK, Peppin JD, White D, Loge F, Call DR (2003) Variation in biofilm formation among strains of Listeria mocytogenes. Appl Environ Microbiol 69:7336–7342
Burall LS, Laksanalamai P, Datta AR (2012) Listeria monocytogenes mutants with altered growth phenotypes at refrigeration temperature and high salt concentrations. Appl Environ Microbiol 78:1265–1272
Chan YC, Boor KJ, Wiedmann M (2007) SigmaB-dependent and SigmaB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl Environ Microbiol 73:6019–6029
Czuprynski CJ (2005) Listeria monocytogenes: silage, sandwiches and science. Anim Health Res Rev 6:211–217
Daines DA, Bothwell M, Furrer J et al (2005) Haemophilus influenzae luxS mutants form a biofilm and have increased virulence. Microb Pathog 39:87–89
Folsom JP, Siragusa GR, Frank JF (2006) Formation of biofilm at different nutrient levels by various genotypes of Listeria monocytogenes. J Food Prot 69:826–834
Garmyn D, Augagneur Y, Gal L, Vivant A-L, Piveteau P (2012) Listeria monocytogenes differential transcriptome analysis reveals temperature-dependent Agr regulation and suggests overlaps with other regulons. PLoS One 7:e43154
Gründling A, Burrack LS, Bouwer HG, Higgins DE (2004) Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc Natl Acad Sci USA 101:12318–12323
Gueriri I, Cyncynatus C, Dubrac S, Arana AT, Dussurget O, Msadek T (2008) The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiology 154:2251–2264
Habimana O, Meyrand M, Meylheuc T, Kulakauskas S, Briandet R (2009) Genetic features of resident biofilms determine attachment of Listeria monocytogenes. Appl Environ Microbiol 75:7814–7821
Harvey J, Keenan KP, Gilmour A (2007) Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol 24:380–392
Hofer E, dos Reis CMF, Hofer CB (2006) Sorovares de Listeria monocytogenes e espécies relacionadas, isoladas de material clínico humano. Rev Soc Bras Med Trop 39:32–37
Ivy RA, Wiedmann M, Boor KJ (2012) Listeria monocytogenes grown at 7 °C shows reduced acid survival and an altered transcriptional response to acid shock compared to L. monocytogenes grown at 37 °C. Appl Environ Microbiol 78:3824–3836
Kamp HD, Higgins DE (2011) A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog. doi: 10.1371/journal.ppat.1002153 doi:10.1371%2Fjournal.ppat.1002153#pmc_ext
Kathariou S (2002) Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Prot 65:1811–1829
Larsen MH, Koch AG, Ingmer H (2010a) Listeria monocytogenes efficiently invades Caco-2 cells after low-temperature storage in broth and on deli meat. Foodborne Pathog Dis 7:1013–1018
Larsen MH, Leisner JJ, Ingmer H (2010b) The chitinolytic activity of Listeria monocytogenes EGD is regulated by carbohydrates but also by the virulence regulator PrfA. Appl Environ Microbiol 76:6470–6476
Lemon KP, Freitag NE, Kolter R (2010) The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes. J Bacteriol 192:3969–3976
Liu D (2006) Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J Med Microbiol 55:645–659
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25:402–408
McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ (2003) LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol 185:274–284
Mello JF, Einsfeldt K, Frazzon APG, Costa M, Frazzon J (2008) Molecular analysis of the iap gene of Listeria monocytogenes isolated from cheeses in Rio Grande do Sul, Brazil. Braz J Microbiol 39:169–172
Moretro T, Langsrud S (2004) Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms 1:107–121
Murray EJ, Kiley TB, Stanley-Wall NR (2009) A pivotal role for the response regulator DegU in controlling multicellular behavior. Microbiology 155:1–8
Nes F, Riboldi GP, Frazzon APG, D’Azevedo P, Frazzon J (2010) Antimicrobial resistance and investigation of the molecular epidemiology of Listeria monocytogenes in dairy products. Rev Soc Bras Med Trop 43:382–385
Neuhaus K, Satorhelyi P, Schauer K, Scherer S, Fuchs TM (2013) Acid shock of Listeria monocytogenes at low environmental temperatures induces prfA, epithelial cell invasion, and lethality towards Caenorhabditis elegans. BMC Genomics 14:285. doi:10.1186/1471-2164-14-285
O’Neil HS, Marquis H (2006) Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect Immun 74:6675–6681
Ollinger J, Bowen B, Wiedmann M, Boor KJ, Bergholz TM (2009) Listeria monocytogenes SigmaB modulates PrfA-mediated virulence factor expression. Infect Immun 77:2113–2124
Pan Y, Breidt F Jr, Gorski L (2010) Synergistic effect of sodium chloride, glucose, and temperature on biofilm formation by Listeria monocytogenes serotype 1/2a and 4b strains. Appl Environ Microbiol 76:1433–1441
Renzoni A, Klarsfeld A, Dramsi S, Cossart P (1997) Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect Immun 65:1515–1518
Riboldi GP, Mattos EP, Frazzon J (2013) Biogenesis of [Fe-S] Cluster in Firmicutes: an unexploited field of investigation. Antonie Van Leeuwenhoek 104:283–300
Rieu A, Briandet R, Habimana O, Garmyn D, Guzzo J, Piveteau P (2008) Listeria monocytogenes EGD-e biofilms: no mushrooms but a network of knitted chains. Appl Environ Microbiol 74:4491–4497
Salter MG, Conlon HE (2007) Extraction of plant RNA. Methods Mol Biol 362:309–314
Sela S, Frank S, Belausov E, Pinto R (2006) A mutation in the luxS gene influences Listeria monocytogenes biofilm formation. Appl Environ Microbiol 72:5653–5658
Shin JH, Brody MS, Price CW (2010) Physical and antibiotic stresses require activation of the RsbU phosphatase to induce the general stress response in Listeria monocytogenes. Microbiology 156:2660–2669
Suo Y, Huang Y, Liu Y, Shi C, Shi X (2012) The expression of Superoxide Dismutase (SOD) and a Putative ABC Transporter Permease is inversely correlated during biofilm formation in Listeria monocytogenes 4b G. PLoS One. doi:10.1371/journal.pone.0048467
Trachoo N (2003) Biofilms and the food industry. Songklanakarin J Sci Technol 25:807–815
Van der Veen S, Abee T (2010) Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl Environ Microbiol 76:7854–7860
Williams T, Bauer S, Beier D, Kuhn M (2005) Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect Immun 73:3152–3159
Acknowledgements
We would like to thank Prof. Giancarlo Pasquali from UFRGS and Prof. Pedro D’Azevedo from UFCSPA for laboratorial and technical support. A special thanks for Eduardo Preusser de Mattos for english spelling support. This work was supported by Brazilian Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (J.F grant #300912/2012-9; #473181/2013-4), and the Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior (CAPES) of Brazil.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
Interaction between PrfA (lmo0200: listeriolysin positive regulatory protein; positively regulates expression of listeriolysin, of 1-phosphadidylinositol phosphodiesterase PI-PLC, and other virulence factors) and other virulence genes of Listeria monocytogenes EGD (STRING 9.05; available in http://string-db.org/) (DOC 78 kb)
Supplemental Table 1
Sequences of primers tested for qPCR data normalization, with respective sizes of the amplified fragments (DOC 35 kb)
Supplemental Table 2
Functions of proteins encoded by genes analyzed in the present study (DOC 34 kb)
Rights and permissions
About this article
Cite this article
Pieta, L., Garcia, F.B., Riboldi, G.P. et al. Transcriptional analysis of genes related to biofilm formation, stress-response, and virulence in Listeria monocytogenes strains grown at different temperatures. Ann Microbiol 64, 1707–1714 (2014). https://doi.org/10.1007/s13213-014-0814-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-014-0814-2