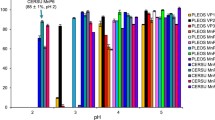
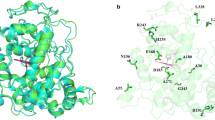
Abstract
A ligninolytic peroxidase called versatile peroxidase, VP, (EC 1.11.1.16) is an iron-containing metalloenzyme. The most distinctive feature of this enzyme is its composite molecular framework, which combines lignin peroxidase’s capacity to oxidize compounds with high-redox potential with manganese peroxidase’s capacity to oxidize Mn2+ to Mn3+. In this study, we have extracted amino acid sequences from the Citrus sinensis source and subjected them to various computation tools to visualize the insight secondary and 3D structure, physicochemical properties, and validation of the structure which have not been studied so far to further investigate the catalytic efficiency and effectiveness of VP. The binding energies of HEME and HEME C (HEC) ligands with produced PDB (6rqf.1. A) have been also assessed, analyzed, and confirmed utilizing AutoDock. Binding energies were calculated using the AutoDock and validated by MD simulation using SCHRODINGER DESMOND. Most stable confirmation was achieved through a protein–ligand interaction study. Bio-technological use of VP in the biotransformation of β-naphthol has also been studied. The findings in the current study will have a substantial impact on proteomics, biochemistry, biotechnology, and possible uses of versatile peroxidase in the bio-remediation of different toxic organic compounds.












Similar content being viewed by others
Data availability
Data will be provided on request to corresponding author.
Abbreviations
- VP:
-
Versatile peroxidase
- LiP:
-
Lignin peroxidase
- MnP:
-
Manganese peroxidase
- WRF:
-
White rot fungus
- NCBI:
-
National Center for Biotechnology Information
- BLAST:
-
The Basic Local Alignment Search Tool
- FASTA:
-
Fast alignment
- pI:
-
Isoelectric point
- SOPMA:
-
Self-optimized prediction method with alignment
- SAVESv6.0:
-
Structure validation server
- PLIP:
-
Protein–ligand interaction profiler
- STRING:
-
Search tool for the retrieval of interacting genes/proteins
- Hh:
-
Alpha helix
- Ee:
-
Extended strand
- Tt:
-
Beta-turn
- Cc:
-
Random coil
- RMSD:
-
Root-mean-square deviation
- RMSF:
-
Root-mean-square fluctuation
- MMGBSA:
-
Molecular mechanics with generalized born and surface area solvation
References
Apweiler R, Bairoch A, Wu CH et al (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32:D115–D119
Artimo P, Jonnalagedda M, Arnold K et al (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40:W597–W603
Barber-Zucker S, Mindel V, Garcia-Ruiz E et al (2022) Stable and functionally diverse versatile peroxidases designed directly from sequences. J Am Chem Soc 144:3564–3571
Basumatary D, Saikia S, Yadav HS, Yadav M (2023) In silico analysis of peroxidase from Luffa acutangula. 3 Biotech 13:25
Bathula R, Lanka G, Muddagoni N et al (2019) Identification of potential Aurora kinase-C protein inhibitors: an amalgamation of energy minimization, virtual screening, prime MMGBSA and AutoDock. J Biomol Struct Dyn 2:2
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350
Bharadwaj S, Rao AK, Dwivedi VD et al (2021) Structure-based screening and validation of bioactive compounds as Zika virus methyltransferase (MTase) inhibitors through first-principle density functional theory, classical molecular simulation and QM/MM affinity estimation. J Biomol Struct Dyn 39:2338–2351
Busse N, Wagner D, Kraume M, Czermak P (2013) Reaction kinetics of versatile peroxidase for the degradation of lignin compounds. Am J Biochem Biotechnol 9:365–394
Camacho C, Coulouris G, Avagyan V et al (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:1–9
Camarero S, Sarkar S, Ruiz-Dueñas FJ et al (1999) Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J Biol Chem 274:10324–10330
Chen VB, Arendall WB, Headd JJ et al (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr 66:12–21
Cohen R, Yarden O, Hadar Y (2002) Lignocellulose affects Mn2+ regulation of peroxidase transcript levels in solid-state cultures of Pleurotus ostreatus. Appl Environ Microbiol 68:3156–3158
Forli S, Huey R, Pique ME et al (2016) Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11:905–919
Gasteiger E, Hoogland C, Gattiker A et al (2005) Protein identification and analysis tools on the ExPASy server. Proteom Protoc Handb 2:571–607
Guillén F, Evans CS (1994) Anisaldehyde and veratraldehyde acting as redox cycling agents for H2O2 production by Pleurotus eryngii. Appl Environ Microbiol 60:2811–2817
Hofrichter M (2002) lignin conversion by manganese peroxidase (MnP). Enzyme Microb Technol 30:454–466
Kumar A, Rajendran V, Sethumadhavan R, Purohit R (2012) In silico prediction of a disease-associated STIL mutant and its affect on the recruitment of centromere protein J (CENPJ). FEBS Open Bio 2:285–293
Kumar A, Singh VK, Kayastha AM (2022) Molecular modeling, docking and dynamics studies of fenugreek (Trigonella foenum-graecum) α-amylase. J Biomol Struct Dyn 2:1–16
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Lee KE, Bharadwaj S, Sahoo AK et al (2021) Determination of tyrosinase-cyanidin-3-O-glucoside and (−/+)-catechin binding modes reveal mechanistic differences in tyrosinase inhibition. Sci Rep 11:24494
Lyne PD, Lamb ML, Saeh JC (2006) Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J Med Chem 49:4805–4808
Perez-Boada M, Ruiz-Duenas FJ, Pogni R et al (2005) Versatile peroxidase oxidation of high redox potential aromatic compounds: site-directed mutagenesis, spectroscopic and crystallographic investigation of three long-range electron transfer pathways. J Mol Biol 354:385–402
Pradeep NV, Anupama AK, Pooja J (2012) Categorizing phenomenal features of α-amylase (Bacillus species) using bioinformatic tools. Adv Lif Sci Technol 4:27–31
Pratama MRF, Siswandono S (2020) Number of runs variations on Autodock 4 do not have a significant effect on RMSD from docking results. Pharm Pharmacol 8:476–480
Purohit R, Rajasekaran R, Sudandiradoss C et al (2008) Studies on flexibility and binding affinity of Asp25 of HIV-1 protease mutants. Int J Biol Macromol 42:386–391
Rai N, Yadav M, Yadav HS (2020) Purification and characterization of versatile peroxidase from citrus sinensis leaf extract and its application in green chemistry. Anal Chem Lett 10:524–536
Ramachandran GNT, Sasisekharan V (1968) Conformation of polypeptides and proteins. Adv Protein Chem 23:283–437
Ruiz-Duenas FJ, Morales M, Mate MJ et al (2008) Site-directed mutagenesis of the catalytic tryptophan environment in Pleurotus eryngii versatile peroxidase. Biochemistry 47:1685–1695
Ruiz-Dueñas FJ, Martínez ÁT (2009) Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb Biotechnol 2:164–177
Salame TM, Yarden O, Hadar Y (2010) Pleurotus ostreatus manganese-dependent peroxidase silencing impairs decolourization of Orange II. Microb Biotechnol 3:93–106
Singh R, Bhardwaj VK, Purohit R (2022) Computational targeting of allosteric site of MEK1 by quinoline-based molecules. Cell Biochem Funct 40:481–490
Takio N, Yadav M, Yadav HS (2022) In silico studies on catalases from plant sources. Authorea Prepr 2:2
Weber K, Osborn M (1969) The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem 244:4406–4412
White SH, Wimley WC (1998) Hydrophobic interactions of peptides with membrane interfaces. Biochim Biophys Acta Reviews Biomembr 1376:339–352
Yadav M, Rai N, Yadav HS (2017a) The role of peroxidase in the enzymatic oxidation of phenolic compounds to quinones from Luffa aegyptiaca (gourd) fruit juice. Green Chem Lett Rev 10:154–161
Yadav M, Yadav S, Yadav D, Yadav KDS (2017b) In-silico analysis of manganese peroxidases from different fungal sources. Curr Proteomics 14:201–213
Yadava U, Yadav VK, Yadav RK (2017) Novel anti-tubulin agents from plant and marine origins: Insight from a molecular modeling and dynamics study. RSC Adv 7:15917–15925
Yang J, Yan R, Roy A et al (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8
Acknowledgements
The Department of Chemistry and CRF of NERIST is well-acknowledged for providing the necessary facilities. I am very thankful for the GATE fellowship provided by NERIST
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
The primary, secondary, and peptide structure analyses and modeling were completed and written manuscript by RAH. The data analysis and interpretation, responsibility, and accountability for the contents of the article were done by MY. MD Simulation and MMGBSA energy calculation has been done by UY and his student SN. NR has done the preliminary study of versatile peroxidase enzymes. HSY was credited for the methodology of the communicated work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Research involving human participants and/or animals
The research work does not involve any human or animal participants.
Informed consent
NA.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hoque, R.A., Yadav, M., Yadava, U. et al. Active site determination of novel plant versatile peroxidase extracted from Citrus sinensis and bioconversion of β-naphthol. 3 Biotech 13, 345 (2023). https://doi.org/10.1007/s13205-023-03758-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03758-x