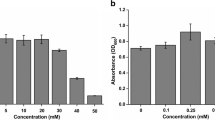
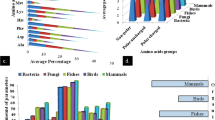
Abstract
Enterobacter cloacae RSC3 isolated from an industrial pesticide site transformed arsenate into arsenite. The arsenate is transported by membrane-bound phosphate transporter and transformed to arsenite by arsenate reductase (arsC). E. cloacae RSC3 produced an arsenate reductase enzyme with a maximum activity of 354 U after 72 h of incubation. Arsenate reductase was found to be active and stable at a wide range of temperatures (20 and 45 °C) and pH (5–10), with maximum activity at 35 °C and pH 7.0. The arsenate reductase protein was further characterised molecularly using different bioinformatics tools. The 3D structure of ArsC protein was predicted by homology modelling and validated by the Ramachandran plot with 91.9% residues in the most favoured region. ArsC protein of E. cloacae RSC3 revealed structural homology with ArsC from PDB ID: 1S3C. The gene ontology results also showed that the ArsC protein had a molecular functionality of the arsenate reductase (glutaredoxin) activity and the biological function of cellular response to DNA damage stimulus. Molecular docking analysis of 3D structures using AutoDock vina-1.5.7 server predicted four ligand binding active site residues at Gln70, Asp68, Leu68, and Leu63. Strong ArsC–arsenate ion interaction was observed with binding energy −1.03 kcal/mol, indicating significant arsenate reductase activity and specificity of ArsC protein. On the basis of molecular dynamics simulation analysis, the RMSD and RMSF values revealed the stability of ArsC protein from E. cloacae RSC3.








Similar content being viewed by others
Availability of data and materials
All data generated and analysed during this study are included in this manuscript and supplementary materials.
References
Ahmad S, Raza K (2023) Identification of 5-nitroindazole as a multitargeted inhibitor for CDK and transferase kinase in lung cancer: a multisampling algorithm-based structural study. Mol Divers. https://doi.org/10.1007/s11030-023-10648-0
Ahmad S, Singh V, Gautam HK, Raza K (2023) Multisampling-based docking reveals Imidazolidinyl urea as a multitargeted inhibitor for lung cancer: an optimisation followed by multi-simulation and in-vitro study. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2209673
Anderson CR, Cook GM (2004) Isolation and characterisation of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr Microbiol 48:341–347. https://doi.org/10.1007/s00284-003-4205-3
Arévalo-Rangel DL, Cárdenas-González JF, Martínez-Juárez VM, Acosta-Rodríguez I (2013) Hexavalent chromate reductase activity in cell free extracts of Penicillium sp. Bioinorg Chem Appl. https://doi.org/10.1155/2013/909412
Bachate SP, Cavalca L, Andreoni V (2009) Arsenic resistant bacteria isolated from agricultural soils of Bangladesh and characterisation of arsenate-reducing strains. J Appl Microbiol 107:145–156. https://doi.org/10.1111/j.1365-2672.2009.04188.x
Bae WC, Lee HK, Choe YC, Jahng DJ, Lee SH, Kim SJ, Jeong BC (2005) Purification and characterisation of NADPH-dependent Cr (VI) reductase from Escherichia coli ATCC 33456. J Microbiol 43:21–27
Banerjee P, Chatterjee A, Jha S, Bhadani NK, Datta PP, Sengupta TK (2022) Biochemical, molecular and in silico characterisation of arsenate reductase from Bacillus thuringiensis KPWP1 tolerant to salt, arsenic and a wide range of pH. Arch Microbiol 204:1–13. https://doi.org/10.1007/s00203-021-02660-5
Beg M, Thakur SC, Meena LS (2018) Structural prediction and mutational analysis of Rv3906c gene of Mycobacterium tuberculosis H37Rv to determine its essentiality in survival. Adv Bioinform 4:1–12. https://doi.org/10.1155/2018/6152014
Bhati R, Sreedharan SM, Rizvi A, Khan MS, Singh R (2022) An insight into efflux-mediated arsenic resistance and biotransformation potential of Enterobacter Cloacae RSC3 from arsenic polluted area. Indian J Microbiol. https://doi.org/10.1007/s12088-022-01028-7
Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, Florida, pp. 11–17
Carugo O, Djinovic-Carugo K (2013) Half a century of Ramachandran plots. Acta Crystallogr 69:1333–1341. https://doi.org/10.1107/S090744491301158X
Chauhan NS, Nain S, Sharma R (2017) Identification of arsenic resistance genes from marine sediment metagenome. IndianJ Microbiol 57:299–306. https://doi.org/10.1007/s12088-017-0658-0
Desai C, Jain K, Madamwar D (2008) Hexavalent chromate reductase activity in cytosolic fractions of Pseudomonas sp. G1DM21 isolated from Cr (VI) contaminated industrial landfill. Process Biochem 43:713–721. https://doi.org/10.1016/j.procbio.2008.02.015
Dong S, Sun J, Mao Z, Wang L, Lu YL, Li J (2020) A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019-nCoV). J Med Virol 92:1542–1548. https://doi.org/10.1002/jmv.25768
Dunivin TK, Yeh SY, Shade A (2019) A global survey of arsenic-related genes in soil microbiomes. BMC Bio 17:1–17. https://doi.org/10.1186/s12915-019-0661-5
Elangovan R, Abhipsa S, Rohit B, Ligy P, Chandraraj K (2006) Reduction of Cr (VI) by a Bacillussp. Biotechnol Lett 28:247–252. https://doi.org/10.1007/s10529-005-5526-z
Forli S et al (2016) Computational protein-ligand docking and virtual drug screening with the autodock suite. Nat Protoc 11(5):905–919
Greenwood JR, Calkins D, Sullivan AP, Shelley JC (2010) Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des 24:591–604
Gupta S, Tewatia P, Misri J, Singh R (2017) Molecular modeling of cloned Bacillus subtilis keratinase and its insinuation in psoriasis treatment using docking studies. Indian J Microbiol 57:485–491. https://doi.org/10.1007/s12088-017-0677-x
Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all-atom protein loop prediction. Proteins 55:351–367
Jorgensen WL, Tirado-Rives J (1988) The OPLS [optimised potentials for liquid simulations] potential functions for proteins, energy minimisations for crystals of cyclic peptides and crambin. J Am Chem Soc 110(6):1657–1666
Kruger MC, Bertin PN, Heipieper HJ, Arsène-Ploetze F (2013) Bacterial metabolism of environmental arsenic mechanisms and biotechnological applications. Appl Microbiol Biotechnol 97:3827–4384. https://doi.org/10.1007/s00253-013-4838-5
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. https://doi.org/10.1021/ci200227
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK - a program to check the stereochemical quality of protein structures. J App Cryst 26:283–291
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486 (PubMed id: 9008363)
Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, Thornton JM (2018) PDBsum: structural summaries of PDB entries. Protein Sci 27(1):129–134
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysistools APIs in 2019. Nucleic Acids Res 47:636–641. https://doi.org/10.1093/nar/gkz268
Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234. https://doi.org/10.1007/s10822-013-9644-8
Mala JGS, Sujatha D, Rose C (2015) Inducible chromate reductase exhibiting extracellular activity in Bacillus methylotrophicus for chromium bioremediation. Microbiol Res 170:235–241. https://doi.org/10.1016/j.micres.2014.06.001
Mora Lagares L, Minovski N, Caballero Alfonso AY, BenfenatiE WS, Culot M, Novič M (2020) Homology modeling of the human p-glycoprotein (Abcb1) and insights into ligand binding through molecular docking studies. Int J Mol Sci 21:4058. https://doi.org/10.3390/ijms21114058
Muller D, Stirn CN, Maier MV (2021) Arsenic removal from highly contaminated groundwater by iron electrocoagulation-investigation of process parameters and iron dosage calculation. Water 13:687. https://doi.org/10.3390/w13050687
Naveed M, Imran K, Mushtaq A, Mumtaz AS, Janjua HA, Khalid N (2018) In silico functional and tumor suppressor role of hypothetical protein PCNXL2 with regulation of the Notch signalling pathway. RSC Adv 8:21414–21430. https://doi.org/10.1039/C8RA00589C
Pande V, Pandey SC, Sati D, Bhatt P, Samant M (2022) Microbial interventions in bioremediation of heavy metal contaminants in agroecosystem. Front Microbiol. https://doi.org/10.3389/fmicb.2022.824084
Park CH, Keyhan M, Wielinga B, Fendorf S, Matin A (2000) Purification to homogeneity and characterisation of a novel Pseudomonas putida chromate reductase. Appl Environ Microbiol 66:1788–1795. https://doi.org/10.1128/AEM.66.5.1788-1795.2000
Rahman M, Hossain M, Saha SK, Rahman S, Sonne C, Kim KH (2021) Homology modeling and probable active site cavity prediction of uncharacterised arsenate reductase in bacterial spp. Appl Biochem Biotechnol 193:1–18. https://doi.org/10.1007/s12010-020-03392-w
Raju NJ (2022) Arsenic in the geo-environment: a review of sources, geochemical processes, toxicity and removal technologies. Environ Res 203:111782. https://doi.org/10.1016/j.envres.2021.111782
Reed JH, Shi Y, Zhu Q, Chakraborty S, Mirts EN, Petrik ID, Lu Y (2017) Manganese and cobalt in the nonheme-metal-binding site of a biosynthetic model of heme-copper oxidase superfamily confer oxidase activity through redox-inactive mechanism. J Am Chem Soc 139:12209–12218. https://doi.org/10.1021/jacs.7b05800
Roy A, Yang J, Zhang Y (2012) COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res 40:W471–W477
Saxena R, Singh R (2010) Metal ion and pH stable protease production using agro-industrial waste. J Ecobiotechnol 2:4
Schrödinger Release 2023-2 (2023a) Prime, Schrödinger, LLC, New York, NY
Schrödinger Release 2023-2 (2023b) Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2023; Impact, Schrödinger, LLC, New York, NY; Prime, Schrödinger, LLC, New York, NY
Schrödinger Release 2023-2 (2023c) Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2023. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY
Selvi MS, Sekar PC, Yuvaraj S, Albert A, Sasikumar EPS, Selvam GS (2015) In silico analysis of arsenate reductase from Enterobacter cloacae BC2 as a potential microorganism for reducing arsenate. J Chem Pharm Res 7:99–105
Sharma C, Salem GEM, Sharma N, Gautam P, Singh R (2019) Thrombolytic potential of novel thiol-dependent fibrinolytic protease from Bacillus cereus RSA1. Biomolecules 10:3. https://doi.org/10.3390/biom10010003
Sharma C, Nigam A, Singh R (2021) Computational-approach understanding the structure-function prophecy of Fibrinolytic Protease RFEA1 from Bacillus cereus RSA1. PeerJ 9:e11570. https://doi.org/10.7717/peerj.11570
Stasi R, Neves HI, Spira B (2019) Phosphate uptake by the phosphonate transport system PhnCDE. BMC Microbiol 19:1–8. https://doi.org/10.1186/s12866-019-1445-3
Vasak M, Schnabl J (2016) Sodium and potassium ions in proteins and enzyme catalysis. The alkali metal ions: their role for life. Springer, Cham, pp 259–290. https://doi.org/10.1007/978-3-319-21756-7_8
Vishnoi N, Singh DP (2014) Biotransformation of arsenic by bacterial strains mediated by oxido-reductase enzyme system. Cell Mol Biol 60:7–14. https://doi.org/10.14715/cmb/2014.60.5.3
Wang C, Liu H, Zhang Y, Zou C, Anthony EJ (2018) Review of arsenic behavior during coal combustion: volatilisation, transformation, emission and removal technologies. Prog Energy Combust Sci 68:1–28. https://doi.org/10.1016/j.pecs.2018.04.001
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303
Yang HC, Fu HL, Lin YF, Rosen BP (2012) Pathways of arsenic uptake and efflux. Curr Top Membr 69:325–358. https://doi.org/10.1016/B978-0-12-394390-3.00012-4
Yin S, Zhang X, Yin H, Zhang X (2022) Current knowledge on molecular mechanisms of microorganism-mediated bioremediation for arsenic contamination: a review. Microbiol Res. https://doi.org/10.1016/j.micres.2022.126990
Zeng XC, Xu Y, Liu Z, Chen X, Wu Y (2022) Comparisons of four As (V)-respiring bacteria from contaminated aquifers: activities to respire soluble As (V) and to reductively mobilise solid-phase As (V). Water Res. https://doi.org/10.1016/j.watres.2022.119097
Zhang Y, Skolnick J (2005) TM-align: a protein structure alignment algorithm based on TM-score. Nucleic Acids Res 33:2302–2309. https://doi.org/10.1093/nar/gki524
Zhang C, Freddolino PL, Zhang Y (2017) COFACTOR: improved protein function prediction by combining structure, sequence, and protein-protein interaction information. Nucleic Acids Res 45:W291-299
Acknowledgements
We extend our sincere thanks to Indian Council of Agricultural Research (AS/22/5/2018-ASR-IV), Government of India, to provide financial support for research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhati, R., Nigam, A., Ahmad, S. et al. Structural–functional analysis and molecular characterization of arsenate reductase from Enterobacter cloacae RSC3 for arsenic biotransformation. 3 Biotech 13, 305 (2023). https://doi.org/10.1007/s13205-023-03730-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03730-9