Abstract
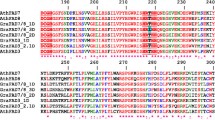
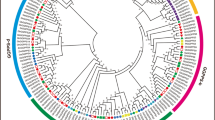
Carotenoid cleavage dioxygenases (CCDs) are a group of enzymes that catalyze the selective oxidative cleavage steps from carotenoids to apocarotenoids, which are essential for the synthesis of biologically important molecules such as retinoids, and the phytohormones abscisic acid (ABA) and strigolactones. In addition, CCDs play important roles in plant biotic and abiotic stress responses. Till now, a comprehensive characterization of the CCD gene family in the economically important crop cotton (Gossypium spp.) is still missing. Here, we performed a genome-wide analysis and identified 33, 31, 16 and 15 CCD genes from two allotetraploid Gossypium species, G. hirsutum and G. barbadense, and two diploid Gossypium species, G. arboreum and G. raimondii, respectively. According to the phylogenetic tree analysis, cotton CCDs are classified as six subgroups including CCD1, CCD4, CCD7, CCD8, nine-cis-epoxycarotenoid dioxygenase (NCED) and zaxinone synthase (ZAS) sub-families. Evolutionary analysis shows that purifying selection dominated the evolution of these genes in G. hirsutum and G. barbadense. Predicted cis-acting elements in 2 kb promoters of CCDs in G. hirsutum are mainly involved in light, stress and hormone responses. The transcriptomic analysis of GhCCDs showed that different GhCCDs displayed diverse expression patterns and were ubiquitously expressed in most tissues; moreover, GhCCDs displayed specific inductions by different abiotic stresses. Quantitative reverse-transcriptional PCR (qRT-PCR) confirmed the induction of GhCCDs by heat stress, salinity, polyethylene glycol (PEG) and ABA application. In summary, the bioinformatics and expression analysis of CCD gene family provide evidence for the involvement in regulating abiotic stresses and useful information for in-depth studies of their biological functions in G. hirsutum.







Similar content being viewed by others
Availability of data and materials
Nine Arabidopsis and 13 rice CCD proteins downloaded from (https://phytozome.jgi.doe.gov/pz/portal.html). All raw RNA-seq data were downloaded from the NCBI Sequence Read Archive (SRA: PRJNA490626).
References
Ablazov A, Mi J, Jamil M, Jia K-P, Wang JY, Feng Q et al (2020) The apocarotenoid zaxinone is a positive regulator of strigolactone and abscisic acid biosynthesis in arabidopsis roots. Front Plant Sci. https://doi.org/10.3389/fpls.2020.00578
Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66:161–186. https://doi.org/10.1146/annurev-arplant-043014-114759
Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P et al (2012) The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 335(6074):1348–1351. https://doi.org/10.1126/science.1218094
Bilas R, Szafran K, Hnatuszkokonka K, Kononowicz AK (2016) Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult 127(2):269–287. https://doi.org/10.1007/s11240-016-1057-7
Bruno M, Beyer P, Al-Babili S (2015) The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch Biochem Biophys 572:126–133. https://doi.org/10.1016/j.abb.2015.02.011
Chen ZJ, Scheffler BE, Dennis ES, Triplett BA, Zhang T, Guo W et al (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol 145(4):1303–1310. https://doi.org/10.1104/pp.107.107672
Chen C, Chen H, He Y, Xia R (2018) TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv. https://doi.org/10.1101/289660
Conant GC, Wolfe KH (2008) Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet 9(12):938–950. https://doi.org/10.1038/nrg2482
Decker EL, Alder A, Hunn S, Ferguson J, Lehtonen MT, Scheler B et al (2017) Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss. Physcomitrella patens New Phytol 216(2):455–468. https://doi.org/10.1111/nph.14506
Endrizzi JE, Turcotte EL, Kohel RJ (1985) "Genetics, cytology, and evolution of gossypium," in Advances in Genetics, (eds) Caspari EW, Scandalios JG (Academic Press), 271–375. https://doi.org/10.1016/S0065-2660(08)60515-5
Fang L, Gong H, Hu Y, Liu C, Zhou B, Huang T et al (2017) Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Bio 18(1):33. https://doi.org/10.1186/s13059-017-1167-5
Felemban A, Braguy J, Zurbriggen MD, Al-Babili S (2019) Apocarotenoids involved in plant development and stress response. Front Plant Sci 10:1168. https://doi.org/10.3389/fpls.2019.01168
Giuliano G, Al-Babili S, Von Lintig J (2003) Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci 8(4):145–149. https://doi.org/10.1016/S1360-1385(03)00053-0
Gonzalez-Jorge S, Ha S-H, Magallanes-Lundback M, Gilliland LU, Zhou A, Lipka AE et al (2013) Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25(12):4812–4826. https://doi.org/10.1105/tpc.113.119677
Hashimot H, Cogdell RJ (2016) Carotenoids and photosynthesis. Subcell Biochem 79:111–139. https://doi.org/10.1007/978-3-319-39126-7_4
Hu B, Jin J, Guo A, Zhang H, Luo J, Gao G (2015) GSDS 2.0 an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y et al (2019) Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51(4):739–748. https://doi.org/10.1038/s41588-019-0371-5
Ilg A, Beyer P, Al-Babili S (2009) Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J 276(3):736–747. https://doi.org/10.1111/j.1742-4658.2008.06820.x
Ilg A, Bruno M, Beyer P, Al-Babili S (2014) Tomato carotenoid cleavage dioxygenases 1A and 1B: Relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Biol 4:584–593. https://doi.org/10.1016/j.fob.2014.06.005
Jia KP, Baz L, Al-Babili S (2017) From Carotenoids to Strigolactones. J Exp Bot 69(9):2189–2204. https://doi.org/10.1093/jxb/erx476
Jia KP, Li C, Bouwmeester HJ, Al-Babili S (2019a) Strigolactone biosynthesis and signal transduction. (eds) Strigolactones - Biology and Applications. 1–45. Springer, Cham. https://doi.org/10.1007/978-3-030-12153-2_1
Jia KP, Dickinson AJ, Mi J, Cui G, Xiao TT, Kharbatia NM et al (2019) Anchorene is a carotenoid-derived regulatory metabolite required for anchor root formation in Arabidopsis. Sci Adv. https://doi.org/10.1126/sciadv.aaw6787
Jia KP, Mi J, Ablazov A, Ali S, Yang Y, Balakrishna A, Berqdar L, Feng Q, Blilou I, Al-Babili S (2021) Iso-anchorene is an endogenous metabolite that inhibits primary root growth in Arabidopsis. Plant J 10(10):15271. https://doi.org/10.1111/tpj.15271
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36. https://doi.org/10.1186/gb-2013-14-4-r36
Kong X, Luo Z, Dong H, Eneji AE, Li W (2016) H2O2 and ABA signaling are responsible for the increased Na+ efflux and water uptake in Gossypium hirsutum L. roots in the non-saline side under non-uniform root zone salinity. J Exp Bot 67(8):2247–2261. https://doi.org/10.1093/jxb/erw026
Ku YS, Sintaha M, Cheung MY, Lam HM (2018) Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci 19(10):3206. https://doi.org/10.3390/ijms19103206
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M (2003) Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J Struct Funct Genomics 3:111–116. https://doi.org/10.1023/A:1022614001371
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327. https://doi.org/10.1093/nar/30.1.325
Li F, Fan G, Wang K, Sun F, Yuan Y, Song G et al (2014) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46(6):567–572. https://doi.org/10.1038/ng.2987
Ma G, Zhang L, Matsuta A, Matsutani K, Yamawaki K, Yahata M et al (2013) Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase 4 in the flavedo of citrus fruit. Plant Physiol 163(2):682–695. https://doi.org/10.1104/pp.113.223297
Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y (2010) Abscisic acid and the control of seed dormancy and germination. Seed Sci Res 20(2):55–67. https://doi.org/10.1017/S0960258510000012
Nisar N, Li L, Lu S, Khin NC, Pogson BJ (2015) Carotenoid metabolism in plants. Mol Plant 8(1):68–82. https://doi.org/10.1016/j.molp.2014.12.007
Ohmiya A (2009) Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol 26(4):351–358. https://doi.org/10.5511/plantbiotechnology.26.351
Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K (2006) Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol 142(3):1193–1201. https://doi.org/10.1104/pp.106.087130
Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14(3):290–295
Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD (2018) HMMER web server: 2018 update. Nucleic Acids Res 46: W200–W204. https://doi.org/10.1093/nar/gky448
Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylides C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109(14):5535–5540. https://doi.org/10.1073/pnas.1115982109
Rodrigo MJ, Alquézar B, Alós E, Medina V, Carmona L, Bruno M et al (2013) A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J Exp Bot 64(14):4461–4478. https://doi.org/10.1093/jxb/ert260
Roy SW, Gilbert W (2005) Complex early genes. Proc Natl Acad Sci USA 102(6):1986–1991. https://doi.org/10.1073/pnas.0408355101
Roy SW, Gilbert W (2006) The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet 7(3):211–221. https://doi.org/10.1038/nrg1807
Roy SW, Penny D (2007) A very high fraction of unique intron positions in the intron-rich diatom thalassiosira pseudonana indicates widespread intron gain. Mol Biol Evol 24(7):1447–1457. https://doi.org/10.1093/molbev/msm048
Rubio-Moraga Á, Rambla J, Fernández-de-Carmen A, Trapero A, Ahrazem O, Orzáez D et al (2014) New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol Biol. https://doi.org/10.1007/s11103-014-0250-5
Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276(5320):1872–1874. https://doi.org/10.1126/science.276.5320.1872
Sexton PD, Gerard CJ (1982) Emergence force of cotton seedlings as influenced by salinity. Agron J 74(4):699–702. https://doi.org/10.2134/agronj1982.00021962007400040025x
Sonnhammer ELL, Ostlund G (2015) InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res 43:234–239. https://doi.org/10.1093/nar/gku1203
Subramanian B, Gao S, Lercher MJ, Hu S, Chen W (2019) Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res 47:270–275. https://doi.org/10.1093/nar/gkz357
Sun Z, Hans J, Walter MH, Matusova R, Beekwilder J, Verstappen FWA et al (2008) Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta 228(5):789. https://doi.org/10.1007/s00425-008-0781-6
Tan B, Joseph LM, Deng W, Liu L, Li Q, Cline K et al (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35(1):44–56. https://doi.org/10.1046/j.1365-313x.2003.01786.x
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. https://doi.org/10.1038/nbt.1621
Voorrips RE (2002) MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78. https://doi.org/10.1093/jhered/93.1.77
Walter MH, Strack D (2011) Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep 28(4):663–692. https://doi.org/10.1039/c0np00036a
Wan XR, Li L (2006) Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem Biophys Res Commun 347(4):1030–1038. https://doi.org/10.1016/j.bbrc.2006.07.026
Wang D, Zhang Y, Zhang Z, Zhu J, Yu J (2010) KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinform 8(1):77–80. https://doi.org/10.1016/S1672-0229(10)60008-3
Wang K, Wang Z, Li F, Ye W, Wang J, Song G et al (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44(10):1098–1103. https://doi.org/10.1038/ng.2371
Wang R, Wang C, Fei Y, Gai J, Zhao T (2013) Genome-wide identification and transcription analysis of soybean carotenoid oxygenase genes during abiotic stress treatments. Mol Biol Rep 40(8):4737–4745. https://doi.org/10.1007/s11033-013-2570-y
Wang Y, Ding G, Gu T, Ding J, Li Y (2017) Bioinformatic and expression analyses on carotenoid dioxygenase genes in fruit development and abiotic stress responses in Fragaria vesca. Mol Genet Genomics 292(4):895–907. https://doi.org/10.1007/s00438-017-1321-5
Wang JY, Haider I, Jamil M, Fiorilli V, Saito Y, Mi J et al (2019) The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat Commun 10(1):810. https://doi.org/10.1038/s41467-019-08461-1
Wang JY, Lin PY, Al-Babili S (2021) On the biosynthesis and evolution of apocarotenoid plant growth regulators. Semin Cell Dev Biol 109: 3–11. https://doi.org/10.1016/j.semcdb.2020.07.007
Wendel JF (1989) New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci USA 86(11):4132–4136. https://doi.org/10.1073/pnas.86.11.4132
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J et al (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33(5):531–537. https://doi.org/10.1038/nbt.3207
Zheng X, Zhu K, Sun Q, Zhang W, Wang X, Cao H et al (2019) Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol Plant 12(9):1294–1307. https://doi.org/10.1016/j.molp.2019.04.014
Zhou Q, Li Q, Li P, Zhang S, Liu C, Jin J et al (2019) Carotenoid cleavage dioxygenases: identification, expression, and evolutionary analysis of this gene family in tobacco. Int J Mol Sci 20(22):5796. https://doi.org/10.3390/ijms20225796
Acknowledgement
We thank Dr. Salim Al-Babili and Dr. Xiongjie Zheng for valuable discussions and critical reading for the manuscript. We thank Dr. Dingding Zhu for technical support.
Funding
This work was funded by the National Natural Science Foundation of China (Grant No. 31900245 and 31770300), the key research and promotion projects (202102110002), Open Fund of State Key Laboratory of Cotton Biology (CB2019A19) and 111 project of China.
Author information
Authors and Affiliations
Contributions
KJ, YM and SZ designed the research. KJ and YG did the qRT-PCR experiments. KJ, YM, SZ, YG, YZ, JG, KL, WF, ZJ and WL analyzed the data with the input of L-SPT. KJ and SZ wrote the manuscript. YM, L-SPT and WL revised the manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest in the publication.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, S., Guo, Y., Zhang, Y. et al. Genome-wide identification, characterization and expression profiles of the CCD gene family in Gossypium species. 3 Biotech 11, 249 (2021). https://doi.org/10.1007/s13205-021-02805-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02805-9