Abstract
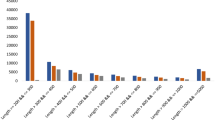
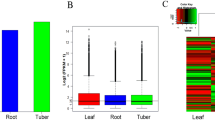
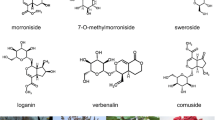
In this study, the putative genes involved in diterpenoid alkaloids biosynthesis in A. vilmorinianum roots were revealed by transcriptome sequencing. 59.39 GB of clean bases and 119,660 unigenes were assembled, of which 69,978 unigenes (58.48%) were annotated. We identified 27 classes of genes (139 candidate genes) involved in the synthesis of diterpenoid alkaloids, including the mevalonate (MVA) pathway, the methylerythritol 4-phosphate (MEP) pathway, the farnesyl diphosphate regulatory pathway, and the diterpenoid scaffold synthetic pathway. 12 CYP450 genes were identified. We found that hydroxymethylglutaryl-CoA reductase was the key enzyme in MVA metabolism, which was regulated by miR6300. Transcription factors, such as bHLH, AP2/EREBP, and MYB, used to synthesize the diterpenes were analyzed.



Similar content being viewed by others
References
Banerjee AB, Hamberger B (2017) P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem Rev 17(1):81–111. https://doi.org/10.1007/s11101-017-9530-4
Botella-Pavía P, Besumbes Ó, Phillips MA, Carretero-Paulet L, Boronat A, Rodríguez-Concepción M (2004) Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J 40(2):188–199. https://doi.org/10.1111/j.1365-313x.2004.02198.x
Carretero-Paulet L, Cairó A, Botella-Pavía P, Besumbes O, Campos N, Boronat A, Rodríguez-Concepción M (2006) Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol 62(4–5):683–695. https://doi.org/10.1007/s11103-006-9051-9
Cherney EC, Baran PS (2011) Terpenoid-alkaloids: Their biosynthetic twist of fate and total synthesis. Isr J Chem 51:391–405. https://doi.org/10.1002/ijch.201100005
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of miRNA genes. Plant Cell 23(2):431–442. https://doi.org/10.1105/tpc.110.082784
Devkota KP, Sewald N (2013) Terpenoid alkaloids derived by amination reaction. In: Ramawat KG, Mérillon JM (eds) Natural products: phytochemistry, botany and metabolism of alkaloids, phenolics and terpenes. Springer, Berlin/Heidelberg, pp 923–951
Dudareva N, Andersson S, Orlova I, Gatto N, Reichelt M, Rhodes D, Boland W, Gershenzon J (2005) The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. PNAS 102(3):933–938
Enfissi EM, Fraser PD, Lois LM, Boronat A, Schuch W, Bramley PM (2005) Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotech J 3(1):17–27. https://doi.org/10.1111/j.1467-7652.2004.00091.x
Gao W, Hillwig ML, Huang LQ, Cui GH, Wang JQ, Bin Y, Peters RJ (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 11:5170–5173. https://doi.org/10.1021/ol902051v
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng QD, Chen ZH, Mauceli E, Hacohen N, Gnirke A, Rhind N, Palma FD, Birren BW, Nusbaum C, Kerstin LT, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech 29(7):644–652. https://doi.org/10.1038/nbt.1883
Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, Rohmer M, Bach TJ (2003) Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J Biol Chem 278(29):26666–26676. https://doi.org/10.1074/jbc.m302526200
Huang J, Liang XM, Xuan YK, Geng CY, Li YX, Lu HR, Qu SF, Mei XL, Chen HB, Yu T, Sun N, Rao JH, Wang JH, Zhang WW, Chen Y, Liao S, Jiang H, Liu X, Yang ZP, Mu F, Gao SX (2017) A reference human genome dataset of the BGISEQ-500 sequencer. GigaScience 6(5):1–9. https://doi.org/10.1093/gigascience/gix024
Jennewein S, Rithner CD, Williams RM, Croteau RB (2001) Taxol biosynthesis: taxane 13-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 98(24):13595–13600. https://doi.org/10.1073/pnas.251539398
Kai GY, Liao P, Zhang T, Zhou W, Wang J, Xu H, Yuanyun Liu YY, Zhang L (2010) Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotech Bioproc Eng 15:236–245. https://doi.org/10.1007/s12257-009-0123-y
Kai GY, Li SS, Wang W, Lu Y, Wang J, Liao P, Cui LJ (2012) Molecular cloning and expression analysis of a gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Camptotheca acuminata. Rus J Plant Physiol 60(1):131–138. https://doi.org/10.1134/s102144371206009x
Lange BM, Ahkami A (2013) Metabolic engineering of plant monoterpenes, sesquiterpenes and diterpenes-current status and future opportunities. Plant Biotech J 11(2):169–196. https://doi.org/10.1111/pbi.12022
Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange BM (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci 100(11):6866–6871
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1):323. https://doi.org/10.1186/1471-2105-12-323
Li Y, Luo HM, Sun C, Song JY, Sun YZ, Wu Q, Wang N, Yao H, Steinmetz A, Chen SL (2010) EST analysis reveals putative genes involved in glycyrrhizin biosynthesis. BMC Genomics 11:268. https://doi.org/10.1186/1471-2164-11-268
Li CF, Zhu YJ, Guo X, Sun C, Luo HM, Song JY, Li Y, Wang LZ, Qian J, Shilin Chen SL (2013) Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng C. A. Meyer. BMC Genomics 14:245. https://doi.org/10.1186/1471-2164-14-245
Li Q, Guo LN, Zheng J, Ma SC (2016) Reaserch progress of medicinal genus Aconitum. Chin J Pharm Anal 36(7):1129–1149
Liao P, Hemmerlin A, Bach TJ, Chye ML (2016) The potential of the mevalonate pathway for enhanced isoprenoid production. Biotech Adv 34(5):697–713. https://doi.org/10.1016/j.biotechadv.2016.03.005
Liao WF, Zhao SY, Zhang M, Dong K, Chen Y, Fu CH, Yu LJ (2017) Transcriptome assembly and systematic identification of novel cytochrome P450s in Taxus chinensis. Front Plant Sci 8:1468. https://doi.org/10.3389/fpls.2017.01468
Liu MM, Zhu JH, Wu SB, Wang CK, Guo XY, Wu JW, Zhou MQ (2018) De novo assembly and analysis of the Artemisia argyi transcriptome and identification of genes involved in terpenoid biosynthesis. Sci Rep 8(1):5824. https://doi.org/10.1038/s41598-018-24201-9
Liu XM, Wang XH, Chen ZX, Ye JB, Liao YL, Zhang WW, Chang J (2019) De novo assembly and comparative transcriptome analysis: novel insights into terpenoid biosynthesis in Chamaemelum nobile L. Plant Cell Rep 38(1):101–116. https://doi.org/10.1007/s00299-018-2352-z
Morris W, Ducreux L, Hedden P, Millam S, Taylor M (2006) Overexpression of a bacterial 1-deoxy-D-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: Implications for the control of the tuber life cycle. J Exp Bot 57(12):3007–3018
Ng DW-K, Zhang CQ, Miller M, Palmer G, Whiteley M, Tholl D, Chen ZJ (2011) Cis- andtrans-regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell 23(5):1729–1740. https://doi.org/10.1105/tpc.111.083915
Pal T, Malhotra N, Chanumolu SK, Chauhan RS (2015) Next-generation sequencing (NGS) transcriptomes reveal association of multiple genes and pathways contributing to secondary metabolites accumulation in tuberous roots of Aconitum heterophyllum Wall. Planta 242(1):239–258. https://doi.org/10.1007/s00425-015-2304-6
Pateraki I, Heskes A, Hamberger B (2015) Cytochromes P450 for terpene functionalisation and metabolic engineering. Adv Biochem Eng Biotechnol 148:107–139. https://doi.org/10.1007/10-2014-301
Pertea G, Huang XQ, Liang F, Antonescu V, Sultana R, Karamycheva S, Karamycheva S, Lee YD, White J, Cheung F, Parvizi B, Tsai J, Quackenbush J (2003) TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19(5):651–652. https://doi.org/10.1093/bioinformatics/btg034
Rai M, Rai A, Kawano N, Yoshimatsu K, Takahashi H, Suzuki H, Kawahara N, Saito K, Yamazaki M (2017) De novo RNA sequencing and expression analysis of Aconitum carmichaelii to analyze key genes involved in the biosynthesis of diterpene alkaloids. Molecules 22(12):2155. https://doi.org/10.3390/molecules22122155
Roberts SC (2007) Production and engineering of terpenoids in plant cell culture. Nature Chem Biol 3(7):387–395. https://doi.org/10.1038/nchembio.2007.8
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Upadhyay AK, Chacko AR, Gandhimathi A, Ghosh P, Harini K, Joseph AP, Sowdhamini R (2015) Genome sequencing of herb Tulsi (Ocimum tenuiflorum) unravels key genes behind its strong medicinal properties. BMC Plant Biol 15:212. https://doi.org/10.1186/s12870-015-0562-x
Xu Y, Qin B, Cao HY, Zhang XN, Liu H, You Y (2017) Simultaneous determination of yunaconitine and bulleyaconitine A in Aconitum vilmorinianum by HPLC. J Yunnan Trad Chin Med 38(9):70–72
Yang CR, Hao XJ, Wang DZ, Zhou J (1981) The alkaloids of Aconitum vilmorrianum Kom. I: the structures of valmorrianine A and C. Acta Chim Sinica 39(2):147–152
Yang L, Ding GH, Lin HY, Cheng HN, Kong Y, Wei YK, Fang X, Liu RY, Wang LG, Chen XY, Yang CQ (2013) Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS ONE 8(11):e80464. https://doi.org/10.1371/journal.pone.0080464
Yuan Y, Song LP, Li MH, Liu GM, Chu YN, Ma LY, Zhou YY, Wang X, Gao W, Qin SS, Yu J, Wang XM, Luqi Huang LQ (2012) Genetic variation and metabolic pathway intricacy govern the active compound content and quality of the Chinese medicinal plant Lonicera japonica Thunb. BMC Genomics 13:195. https://doi.org/10.1186/1471-2164-13-195
Yuan Y, Yu M, Jia Z, Song X, LiangY ZJ (2018) Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genomics 19:978. https://doi.org/10.1186/s12864-018-5305-6
Zhang B, Zhang W, En R, Li WZ, Segraves KA, Yang XK, Xue HJ (2016) Comparative transcriptome analysis of chemosensory genes in two sister leaf beetles provides insights into chemosensory speciation. Insect Biochem Mol Biol 79:108–118. https://doi.org/10.1016/j.ibmb.2016.11.001
Zhao D, Shen Y, Shi YN, Shi XQ, Qiao Q, Zi SH, Zhao EQ, Yu DQ, Kennelly EJ (2018a) Probing the transcriptome of Aconitum carmichaelii reveals the candidate genes associated with the biosynthesis of the toxic aconitine-type C19-diterpenoid alkaloids. Phytochemistry 152:113–124. https://doi.org/10.1016/j.phytochem.2018.04.022
Zhao Q, Li R, Zhang Y, Huang KJ, Wang WG, Li J (2018b) Transcriptome analysis reveals in vitro-cultured regeneration bulbs as a promising source for targeted Fritillaria cirrhosa steroidal alkaloid biosynthesis. 3 Biotech 8:191. https://doi.org/10.1007/s13205-018-1218-y
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (No. 31560351; No. 31760349).
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. Y-GL conceived and designed the experiments, performed the experiments, wrote the paper, read and approved the final manuscript. F-JM contributed reagents, materials, analysis tools, read and approved the final manuscript. K-ZL conceived and designed the experiment, read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, YG., Mou, FJ. & Li, KZ. De novo RNA sequencing and analysis reveal the putative genes involved in diterpenoid biosynthesis in Aconitum vilmorinianum roots. 3 Biotech 11, 96 (2021). https://doi.org/10.1007/s13205-021-02646-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02646-6