Abstract
The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) has been established now to be a deadly disease afflicting the whole world with worst consequences on healthcare, economy and day-to-day life activities. Being a communicable disease, which is highly pathogenic in humans, causing cough, throat infection, breathing problems, high fever, muscle pain, and may lead to death in some cases especially those having other comorbid conditions such as heart or kidney problems, and diabetes. Finding an appropriate drug and vaccine candidate against coronavirus disease (COVID-19) remains an ultimate and immediate goal for the global scientific community. Based on previous studies in the literature on SARS-CoV infection, there are a number of drugs that may inhibit the replication of SARS-CoV-2 and its infection. Such drugs comprise of inhibitors of Angiotensin-Converting Enzyme 2 (ACE2), transmembrane Serine Protease 2 (TMPRSS2), nonstructural protein 3C-like protease, nonstructural RNA-dependent RNA polymerase (RdRp) and many more. The antiviral drugs such as chloroquine and hydroxychloroquine, lopinavir and ritonavir as inhibitors for HIV protease, nucleotide analogue remdesivir, and broad-spectrum antiviral drugs are available to treat the SARS-CoV-2-infected patients. Therefore, this review article is planned to gain insight into the mechanism for blocking the entry of SARS-CoV-2, its validation, other inhibition mechanisms, and development of therapeutic drugs and vaccines against SARS-CoV-2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
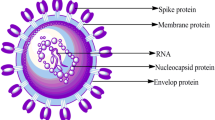
The year 2020 witnessed an outbreak of devastating and pandemic coronavirus disease, COVID-19 which is caused by SARS-CoV-2. The very first case of mortality due to COVID-19 was reported in late December 2019 in the Wuhan city of China (Saxena 2020; Singh and Florez 2020). Coronavirus has the largest single-stranded positive RNA genome which is packed in the enveloped nucleocapsid protein. It is spherical or pleomorphic in structure with spike- or crown-like projections of glycoproteins with 80–120 nm in diameter on its surface, 6–10 open-reading frames, and 26.2–31.7 kb in size (Yang et al. 2006; Guo et al. 2008; Prajapat et al. 2020; Sood et al. 2020). Till now, there are seven important strains of human coronavirus (Chang et al. 2016), whose conserved genes (McBride et al. 2014) and structural components (Hilgenfeld 2014) have been depicted in Fig. 1. The main hypothesized reservoir of SARS-CoV-2 are considered to be bats which transmit the virus to human beings with symptoms such as common cold, respiratory tract infections, cough, fever bronchiolitis and many more (Saif 2004). During the emergence of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) in Guangdong province of China in 2003 (http://www.who.int/csr/sars/country/table2004_04_21/en/) and the Middle East respiratory syndrome (MERS-CoV) in the year 2012 in Saudi Arabia, they were declared as emerging viruses because of being adaptive to the changing environments. However, the COVID-19 virus was declared as public emergency on 30 January 2020 by World Health Organization (WHO) because this virus mutated rapidly and its recombination frequency was very high, which further gave rise to newer strains with new virulence characteristics. Therefore, to protect the world, special precautions of social distancing, wearing of masks, sanitization of containment zones, quarantine and lockdowns were adopted by the respective Governments and people (Gennaro et al. 2020; Upadhyay et al. 2020). The onset of SARS-CoV-19 in Wuhan and the status of infected people worldwide have been shown in Fig. 2. The increase in the number of patients in coronavirus with symptomatic and asymptomatic conditions and lack of a proper vaccine or drug available for the treatment of patients, have led to an increase in death rate. Therefore, immediate approaches are required to study and design novel antiviral drugs to combat with COVID-19 (Abdul-Rahman et al. 2020; Down et al. 2020; Jin et al. 2020; Bhatia et al. 2020). To achieve this, various pharmaceutical companies, researchers, drug discovery labs and organizations have explored the identification and evaluation of the target and along with the underlying mechanism (Alexander et al. 2020a, b; Jeon et al. 2020). Across the world, various therapies, treatments, and clinical trials of vaccines are being carried out by various firms including Moderna and NIAID, BioNTech and Pfizer, Inovio Pharmaceuticals, University of Oxford, AstraZeneca, CanSino Biologics, Wuhan Institute of Biological Products, Beijing Institute of Biological Products, Sinopharm, Sinovac, Institute of Medical Biology and Chinese Academy of Medical Sciences and Novavax. (Talwar et al. 2020; Mullard 2020). In this article herein, we review the potential drug targets and the putative mechanisms thereof against SARS-CoV-2. Moreover, the review also includes the RNA synthesis inhibitors, antiviral therapies, inhibition of viral endocytosis and ACE2 receptors, antibiotics, inhibition of inflammatory mechanisms mediated by different cytokines and phagocytosis mechanism as well.
Chemistry of major anti-COVID-19 drugs
Since the beginning of year 2020, COVID-19 has created havoc in the world. It is of paramount importance to develop a vaccine against this virus which is a time-consuming process and need clinical trials (Jiang et al. 2020). In 2020, Chang et al. recommended drugs which have been used against viral infections which were already approved by US Food and Drug Administration (FDA). Various investigations have been carried out for the treatment of COVID-19 with antiviral compounds which have been proven to inhibit the COVID-19 infection. The proposed drugs which are reported to work against the virus infection with their molecular formula and mechanism of action, have been tabulated in Table 1. The innate immune system of humans plays an important role in defense against SARS-CoV-19 replication. With the help of interferons, the immune response and the binding of virus to the Angiotensin-Converting Enzyme 2 (ACE2) increase (Omrani et al. 2014). Coronavirus can be blocked by inhibiting the virus self-assembly through structural proteins, virus association with human cells, inhibiting the replication process of virus by blocking several enzymes and the viral transcription factors (Saikatendu et al. 2005). There were three main criteria and strategies based on antiviral compounds which were adopted by scientists and researchers to treat COVID-19 (Huang et al. 2019). The first input was to use the drugs associated with cyclophilin inhibitors, interferons and ribavirin which are associated with pneumonia-like symptoms, hence approved for the virus treatment. The major disadvantage of these drugs is that they are broad spectrum and killing the virus in a targeted manner still requires a more in-depth research. The second input was to use the existing literature, molecular databases and to find the new molecules with therapeutic potential which are useful and effective against COVID-19 or coronavirus. The anti-HIV drugs such as ritonavir and lopinavir were discovered as potential therapeutics with high-throughput techniques which further help in the effective functioning of the new molecules (Chen et al. 2014; Mummed 2020). The last input was based on the coronavirus genetic and pathological features. The 3-D structures of drugs used for the treatment of the Coronavirus-19 have been shown in Fig. 3 which are retrieved from PubChem database (Kim et al. 2016). It was supposed that development of drugs through this method will be more efficient and powerful against the virus. But the method was found to be time consuming which could take even more than 10 years to develop the drug after passing all phases of the clinical trials (Omrani et al. 2014; Mummed 2020).
Mechanistic insight
The new therapies for treating the coronavirus are known to act by inhibiting the viral targets and by controlling the infection caused by nucleotide analogues (Raj et al. 2020; Chandel et al. 2020a). The surface glycoproteins of viruses (Influenza, Ebola, SARS, and MERS) require proteolytic cleavage by a host cell protease. The coronavirus-2 enters the host cells by ligating the spike proteins of the outer membrane of virus with ACE2 receptor as host cell which is further primed by Transmembrane serine protease 2 (TMPRSS2 protease). The replication of the virus and assembly can be blocked by antiviral drugs which target protease 3-chymotrypsin-like cysteine protease (3Clpro) and RNA-dependent RNA polymerase (RdRp) (Bertram et al. 2012; Zhou et al. 2015; Yamaya et al. 2015).
Inhibition of viral endocytosis
Endocytosis is a fundamental cellular process through which extracellular material is brought into the cell interior. Classically, it occurs via the regulation of two different modules—pinocytosis (cell drinking) and phagocytosis (cell eating) depending upon the type of molecules approaching towards the cell surface. The foreign molecules (viruses, bacteria, etc.) use endocytosis process to enter inside the cell. Nowadays, it is the utmost need of biologists and researchers to understand the endocytosis mechanism to stop the pathogen particles to enter inside the cell. However, it is not clear until date how this process is regulated at the molecular level. Classically, pinocytosis involves the uptake of liquid particles while phagocytosis, an actin-dependent course involves the internalization of viruses or bacterial particles to the cell (Dutta and Donaldson 2012). In addition, pinocytosis can be sub-categorized into clathrin-mediated endocytosis (CME) and clathrin-coated independent endocytosis (CIE).
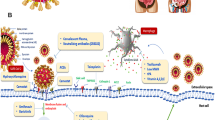
Cell biologists are now trying to block the entry of SARS-CoV-2 inside the cell via inhibiting the endocytosis process, a novel therapeutic strategy to make the vaccine for curing Covid-19 patients. Endocytosis inhibitors could be chemically and genetically originated consisting of selective pharmacologic agents, molecular agents (inhibitors of selective cell-surface proteins and ribonucleic acid). In the current scenario, therapeutic targets are being tried to design specifically to block the attachment of SARS-CoV-2 with cell-surface protein and endocytosis process (Fig. 4). The proposed drugs for Covid-19 such as Nafamostat and Camostat block the fusion of SARS-CoV-2 to cell-surface protein (TMPRSS2). Chloroquinone blocks the ACE2 protein–SARS-CoV-2 fusion. Similarly, the other drug Imatinib inhibits the endocytosis process. Baricitinib drug also inhibits the clathrin-mediated endocytosis by blocking the numb-associated kinase (NAK) particularly AAK1 (Stebbing et al. 2020). As per our best knowledge, chemically and genetically designed inhibitors have been utilized in the preparation of therapeutic targets against SARS-CoV-2 virus, listed in Table 2 (Stebbing et al. 2020; Yang and Shen 2020).
Inhibition of ACE2 receptor
ACE2 is mainly present in lungs, kidney, testis, and heart (Donoghue et al. 2009; Ferrario and Varagic 2010; Ohtsuki et al. 2010) and was first discovered in 2000 by Donoghue et al. (2009). It is expressed in non-keratinized squamous epithelium basal layer of nasopharynx, oral and nasal mucosa basal epidermal skin layer with the strongest expression in type II epithelial cells (Cheng et al. 2020). However, ACE2 is not expressed in glomerular endothelial and mesangial cells whereas it is slightly expressed in glomerular tubules. Further B cells, T cells, bone marrow, macrophages, thymus, spleen, Kupffer cells, lymph nodes, and hepatocytes do not depict the presence of ACE2 (Hamming et al. 2004; Santos et al. 2018). ACE2 activity of tissues is higher as compared to its plasma activity (Haber et al. 2014). ACE inhibitors do not inhibit the activity of ACE2 (Donoghue et al. 2009). ACE2 possesses 400 times higher efficiency for angiotensin II (Ang II) than angiotensin I (Ang I) (Vickers et al. 2002) with difference in its activity depending on the sex and age of individuals (Xudong et al. 2006; Soro-Paavonen et al. 2012).
One of the worst symptoms of COVID-19 is acute respiratory distress syndrome (ARDS) which causes increased pulmonary edema and pulmonary vascular permeability. The novel coronavirus for COVID-19 (SARS-CoV-19) invades the human alveolar epithelial cells through ACE2 (Yang and Shen 2020). ACE2 exhibits a protective role in case of ARDS (Kuba et al. 2005; Xu et al. 2020a, b). When COVID-19 binds to ACE2 receptor it activates the transmembrane serine protease 2 (TMPRSS2), the envelope of the virus fuses with the membrane and thus invades the cells (Fig. 5) (Heurich et al. 2014; Hoffmann et al. 2020). ACE2 generates Ang (1–9) by the cleavage of leucine from Ang I and Ang (1–7) by the cleavage of phenylalanine from Ang II (Rossi et al. 2020). The detrimental effects such as fibrosis, inflammation, and increased vascular permeability, caused by Ang II are counteracted by Ang (1–7). Therefore, ARDS is improved by angiotensin-receptor blockers (ARBs) or ACEIs and Ang (1–7) (Kuba et al. 2005; Wösten-van Asperen et al. 2011; Imai et al. 2005). The COVID-19 entry to the lung cells could be prevented by TMPRSS2 inhibitors such as nafamostat mesylate (Yamamoto et al. 2016) and comstat (Hoffmann et al. 2020) or neutralizing antibodies present in SARS convalescent sera.
Since ACE2 was spotted as the receptor for COVID-19, there were two scientifically unsupported articles which contended that ARB- and ACEI-supplemented treatment for COVID-19, was harmful for the patients. This caused distress amongst the public and health officials as more patients began to question the treatment given to them. It was also documented in the Lancet Respiratory Medicine journal, that the cardiac (diabetes and/or hypertension) diseased patients developed higher amounts of ACE2 after ARB and ACEI treatments (Fang et al. 2020). However, upon reviewing the literature, it was found that the higher amount of ACE2 secretion was due to conditions like myocardial infarction (Ishiyama et al. 2004; Burrell et al. 2005; Ocaranza et al. 2006) rather than the treatment with drugs for COVID-19 (Rossi et al. 2020). It was found that the increased levels of ACE2 aided the blocking of 2019-nCoV attachment to the lung cells by jamming its S protein (Fig. 5). This strategy was used in the development of anti-COVID-19 drugs to prevent the lung infection caused by 2019-nCoV (Kruse 2020; Zhang et al. 2020). This hypothesis was spread by various scientific societies such as the Italian Society of Arterial Hypertension, the Italian Society of Cardiology and the European Society of Hypertension (ESH) and encouraged the patients to continue their ARB- and ACEI-based treatments (Greene et al. 2013; Danser et al. 2020; Perico et al. 2020). In fact, it was found in a study that adverse health effects and clinical instability were observed in high-risk patients if the ARB and ACEI-based treatments were withdrawn abruptly (Vaduganathan et al. 2020).
Inhibition of proteolysis via 3CLpro and PLpro
The structure of 2019-nCoV consists of six open-reading frames (ORFs) which code for the synthesis of sub-genomic mRNAs. The frameshift mutations amongst ORF1a and ORF1b are responsible for coding pp1a (486 kDa) and pp1ab (790 kDa) polyproteins (Muramatsu et al. 2016; Rana et al. 2020). These polyproteins are cleaved into functional proteins by papain-like protease (PLpro) and 3-chymotrypsin-like protease (3CLpro), also known as main protease (Mpro) (Ziebuhr et al. 2000). The main function of PLpro is to defend the coronavirus from immune reaction of the host by removing ubiquitin, whereas 3CLpro aids in the synthesis of functional proteins responsible for translation and replication of the virus. Plant-derived products (Akram et al. 2018) or some other natural products (Aanouz et al. 2020; Borkotoky and Banerjee 2020; Wahedi et al. 2020) that inhibit 3CLpro can be used as anti-COVID-19 drugs (Al-Obaidi et al. 2020; Bhardwaj et al. 2020; Kumar et al. 2020). In a study, 32,297 compounds from medicinal plants known for their antiviral properties were screened and it was found that nine of them (Myricitrin, Methyl rosmarinate, 5,7,3′,4′-Tetrahydroxy-2′-(3,3-dimethylallyl) isoflavone, Calceolarioside B, 3,5,7,3′,4′,5′-hexahydroxy flavanone-3-O-beta-d-glucopyranoside, Licoleafol, Myricetin 3-O-beta-d-glucopyranoside, Amaranthin, (2S)-Eriodictyol7-O-(6″-Ogalloyl)-beta-d-glucopyranoside) could be used as the anti-COVID-19 drugs. However, these drugs need to be analyzed in vitro and in vivo, before using them as clinical drugs to treat COVID-19 patients (ul Qamar et al. 2020). Chloroquine and formoterol are also known to inhibit the PLpro (Arya et al. 2020).
Proteases have always been a key component in the synthesis of antiviral drugs such as in case of hepatitis C virus and human immunodeficiency virus (HIV) (De Clercq 2002; Turk 2006; Chandel et al. 2020b). Since 3CLpro and PLpro are essential for 2019-nCoV survival, these can be targeted as anti-COVID drugs (Han et al. 2005). PLpro possesses papain-like cysteine protease characteristics, three self-cleaving sites and deubiquitinating activity (Barretto et al. 2005; Han et al. 2005). It is known to hydrolyze synthetic ubiquitin peptide substrate, polyubiquitin and diubiquitin in vitro (Barretto et al. 2005; Lindner et al. 2005). In a study, 6-thioguanine (6TG) and 6-mercaptopurine (6MP) were found to be slow-binding inhibitors of PLpro in 2019-nCoV (Chou et al. 2008) through allosteric inhibition which change the conformation of active site making it unavailable for binding. Like previous antiviral drug formulations (O'Connor and Roth 2005), these compounds can be optimized as anti-COVID-19 drugs. However, these drugs can be cytotoxic to the patients if administered in higher dosages (Chou et al. 2008).
3CLpro possesses a His41 which behaves as a basic acid–base during proteolysis and an active site with Cys145 (Liu and Zhou 2005). TG-0205221 and chloromethyl ketone (CMK) (Anand et al. 2003) were first-generation 3CLpro inhibitors forming a covalent bond between the reactive atom and Cys145 (Anand et al. 2003; Xue et al. 2008) giving rise to a strong protease interaction. However, these drugs exhibit some adverse side effects such as lower potency, toxicity, and off-target reactions (Ghosh et al. 2010; Tuley and Fast 2018). A strong inhibitory interaction was observed with the binding of Cm-FF-H, a peptide aldehyde inhibitor, to protease of 2019-nCoV. The ligand bound non-covalently to hydrophilic pocket of protease via its electrophilic P1-phenyl alanine residue (Zhu et al. 2011). This inhibitor explicated its inhibitory activity by catalyzing the conversion of trifluoromethyl-b-amino alcohol to four tetra- and triglutamic acid and glutamine peptides (Sydnes et al. 2006). Good to moderate inhibitory activity has been observed using tripeptidic Z-Val-Leu-Ala(pyrrolidone-3-yl)-2-benzothiazole (Hosseini-Zare et al. 2020). As the peptidyl inhibitors possess weaker pharmacokinetics profiles, impermeability to cell membranes and in vivo instability, caution should be taken while considering them as 3CLpro inhibitors.
Inhibition of replication complex via RNA-dependent RNA polymerase
COVID-19 is characterized by a highly conserved genomic organization, unique enzymatic activities, and expression of nonstructural genes by ribosomal frame-shifting (Fehr and Perlman 2015). Mature SARS-CoV-2 virion reproduction depends on the host cellular mechanisms. A plausible therapeutic target to inhibit RNA replication from an RNA template which leads to three potential benefits as depicted in Fig. 6. Even though low-level nonstructural proteins (NSPS) production is still possible inside the cell, but eliminating the viral infection is only possible by targeting the RNA replication of the virus (Astuti and Ysrafil 2020).
The genome of coronavirus includes a 5′ cap structure along with a 3′ poly (A) tail that serves as a mRNA for translation, and this is responsible for translation of the replicase polyproteins. Two-thirds of the genome is formed by the replicase gene which encodes the NSPS. Several accessory proteins have been shown to play a role in viral pathogenesis, but these are not essential for viral replication but are also included in the genome (Sood et al. 2020).
Once the viral genome enters the cytoplasm, the host ribosomes translate the viral positive-sense RNA into 16 NSPS that may be engaged in making sure that the viral RNA is translated efficiently without interference from the host. Some NSPS are vital for the production of mature virions, while the other NSPS develop the replicase-transcriptase complex (RTC) that consists of proteins needed to replicate both genomic and sub-genomic viral RNA through anti-sense RNA intermediates (Ziebuhr 2005). The viral structural proteins enter the endoplasmic reticulum (ER) following viral genomic RNA replication and viral protein translation from sub-genomic RNA. The proteins then follow the secretory pathway and develop the endoplasmic reticulum–Golgi intermediate compartment (ERGIC). Following this formation, the N protein encapsulated viral RNA genomes get incorporated into the ERGIC, and then bud into mature virions which eventually exit the cell via exocytosis and infect other cells, thereby spreading the virus load (Choudhury et al. 2020).
Besides treating the various symptoms of COVID-19, clinicians can opt for targeted treatment for the SARS-CoV-2 by targeting viral-specific processes such as viral replication through RdRp inhibition. This is a better approach as it stops the formation of mature virions and stops the virus from spreading and damaging other host cells (Warren et al. 2016; Siegel et al. 2017).
Antiviral drugs that target the RdRp active sites such as the ASP760 and ASP761 may potentially serve as a therapeutic option. Anti-viral drugs such as Favipiravir are viral-specific, and they inhibit the RdRp and result in the inhibition of SARS-CoV-2. Favipiravir structurally has a carboxamide (C–[O]–NH2) moiety that targets the viral RNA polymerase, and its active form blocks the catalytic domain of the viral RNA-dependent RNA polymerase, and its enzymatic activity. This effectively ends the infectious cycle of SARS-CoV-2. Favipiravir is not toxic to mammalian cells since it does not inhibit RNA or DNA synthesis within mammalian cells (Holshue et al. 2020).
Inhibition of calcineurin NEFT pathways
In humans, transplantation of organs such as heart, lungs, liver and kidneys are being performed more frequently. In response to this transplantation, lifelong dual or triple immunosuppressive therapy is needed to prevent the organ rejection. COVID-19 has become a problem in solid-organ transplant (SOT) patients (Willicombe et al. 2020) and there is no clue to deal with immunosuppression in SOT recipients having COVID-19. Immunosuppressive drugs named as calcineurin inhibitors (CNIs), a calcium–calmodulin-activated serine/threonine-specific phosphatase, an immunosuppressive drug, which acts as an important inhibitor in the prevention of rejection. On the other hand, drugs named as cyclosporin and tacrolimus are helpful in transplantation of medicine in CNIs. (Hage et al. 2020). Tanaka et al. (2013) studied that cyclosporin suppressed the replication process of SARS-CoV-1 which is another common virus of Severe Acute Respiratory Syndrome (SARS). Calcineurin inhibitors (CNIs) such as cyclosporin and its derivatives such as cyclosporine A-derivatives inhibit the calcineurin pathway by binding to cellular cyclophilins (Cyps) and NL63 is inhibited by cyclosporine A-derivatives (Carbajo-Lozoya et al. 2014; Sanchez-Pernaute et al. 2020). Tacrolimus, another immunosuppressive drug, inhibits calcineurin by suppressing the early phase activation of T cells and expression of cytokines resulting in the suppression of immune-cellular responses, which lead to the prevention of blizzard of cytokines (IL-2, IL-4, TNF-α and IFN-γ) in COVID-19.
Inhibition of de-regulated proinflammatory response in COVID-19
A cytokine is a signaling molecule responsible for the regulation of distinct biological functions via receptors present on the surface of the cell (Bartee and McFadden 2013) while chemokine is a specific cytokine for attracting the cells to the site of inflammation or infection. Cellular infection followed by viral replication leads to the release of proinflammatory cytokines and death of the cell resulting in an amplified inflammatory response due to release of damage-associated molecular pattern (Tay et al. 2020; Siu et al. 2019; Chen et al. 2019). The exact nature of the inflammatory response depends upon the type of virus and the tissue that has been infected (Thomas et al. 2009; Allen et al. 2009). Cytokine Release Syndrome (CRS) or Cytokine storm is a condition of exaggerated release of cytokines after infection by a virus and it is emerging as a leading cause of Multiple Organ Failure (MOF) and Acute Respiratory Distress Syndrome (ARDS) in COVID-19 (Tay et al. 2020).
Severely affected patients have been found to have raised levels of interleukins (IL): (IL)-1β, IL-2, IL-6, IL-7, IL-8, IL-9, IL-10, IL-18, TNF-α, Macrophage Colony-Stimulating Factor (M-CSF), Granulocyte Macrophage-Colony-Stimulating Factor (GM-CSF), Granulocyte-Colony-Stimulating Factor (G-CSF), 10 kD interferon-gamma-induced protein (IP-10), Macrophage Inflammatory Protein 1-α (MIP 1-α) and Monocyte Chemoattractant Protein-1 (MCP-1) in the serum (Huang et al. 2019; Liu et al. 2020; Wang et al. 2020). These studies focused on the potential role of immunosuppressive or immunomodulatory approaches to combat unregulated proinflammatory response.
The COVID-19 infection involves three phases. It starts from an asymptomatic phase in which virus is shredded in high amount in the upper respiratory tract. In the second phase, which is the non-severe symptomatic phase, adaptive immune activation occurs which allows the development of B (generation of specific antibodies) and T cell responses (Fig. 7). In approximately 80% of the patients, this phase marks the end of the disease. Final phase involves the release of high levels of inflammatory cytokines. This phase is also characterized by MOF, progressive fever, hypercoagulability and shock (Berlin et al. 2020). Studies have shown that the inhibition of excessive inflammatory mechanisms contributes in eliminating the viral resistance. Immunomodulatory strategies involved in the treatment of COVID-19 infection targeting certain cytokines are listed in Table 3.
Role of plasma therapy and its mechanism
The convalescent plasma of patients recovered from COVID-19 infection can be used to meet the requirements of antiviral antibodies in newly affected COVID-19 patients. Various beneficial actions of convalescent plasma can, therefore, be availed to help fight against new COVID-19 infection cases. Most viral illnesses manifest a viremia peak within the first week of infection, and it takes about 10–14 days for the host to develop a primary immune response that signals virus clearance. However, viral antibodies contained in convalescent plasma enable viremia suppression, especially if given at an early stage of the disease. Antibody-dependent cellular cytotoxicity (ADCC), complement activation and phagocytosis are also some of the mechanisms that come in effect to clear the virus load. Inflammatory-response suppression and coagulation factors restoration are also plausible mechanisms that help fight against the COVID-19 disease, especially when convalescent plasma is administered early, even to asymptomatic cases (Gunn et al. 2018; Van Erp et al. 2019).
Donor criteria
US FDA has given the approval for the use of COVID-19 convalescent plasma in COVID-19 patients who are at a serious or immediate life-threatening stage of the disease on March 24, 2020. Only certified blood laboratories and technicians should collect plasma with proper approved equipment. The volume of the plasma collected at any one time can be between 200 and 600 ml, with a gap of at least 7 days before another collection from the same donor. Donors eligible to give convalescent plasma are defined as meeting the following criteria: (1) recovered COVID-19 patients that were COVID-19 positive either a positive nasopharyngeal swab at the time of illness, or those who were antibody-positive even if no diagnostic test was done during their illness. (2) A minimum antibody titer of 1:160 should be there in the convalescent plasma. A titer of 1:80 may be accepted only if alternative matching units are unavailable. (3) They should be totally symptom-free for a minimum of 28 days prior to their donation. Exemptions may be considered if the donors are at least symptom-free for 14 days before donation, and they have negative nasopharyngeal swab reports, or negative blood-based molecular diagnostic test results. (4) Female donors must be screened for human leukocyte antigen (HLA) antibodies negativity in case of prior pregnancies; however, male donors are eligible without performing this screening. Transfusion-related acute lung injury (TRALI) is a serious complication that can occur within 6 h of transfusion, and therefore, male donors are preferred, and prior pregnancy and abortion must be ruled out in females before donor consideration. (5) Donors must be eligible generally, with a minimum 14-day interval between full recovery and donation, and consideration of their infection control status and plasmapheresis should be done. Their samples should be negative for HIV, hepatitis B virus (HBV), hepatitis C virus (HCV), syphilis, and locally transmitted infections (Franchini et al. 2020).
Recipient criteria
Convalescent plasma transfusion is generally considered to be safe and without serious adverse effects. However, the risk of TRALI should be kept in mind, and donor selection criteria should be adhered to, meticulously. After getting an informed consent from the patient, convalescent plasma should be given to COVID-19 positive patients who have severe disease symptoms as defined by the following criteria like respiratory rate should be ≥ 30/min whereas blood oxygen saturation must be ≤ 93%. On the other hand, the ratio of partial pressure of arterial oxygen to fraction of inspired oxygen ratio should be less than 300 otherwise, it may lead to life-threatening disease conditions such as respiratory failure, septic shock, or multiple organ dysfunction (Franchini et al. 2020).
Therapeutic role of convalescent plasma
More than 15,521,145 patients have already recovered from COVID-19 globally by 19 September 2020. The convalescent plasma is definitely a powerful life-saving therapeutic option for newly affected COVID-19 patients. In addition, more donors can also be channeled for providing therapy, if they are asymptomatic but antibody-positive. Convalescent plasma administration within 10–11 days after symptoms, may result in a quick rise in lymphocyte counts, diminished CRP levels, and an obvious resolution of lung lesions of CT scans. Results obtained from convalescent plasma administration have been considered to be promising without any serious adverse effects. Convalescent plasma administration should be given as early as possible to severe cases to maximize its efficacy which could certainly alter the morbidity and mortality burden associated with the COVID-19 pandemic (Chang et al. 2020a, b; Chen et al. 2020).
Conclusions
From the past hundreds of years, the SARS virus has been proved to be the greatest worldwide public health crisis. The present century also became the victim of SARS-CoV-2 causing the disease popularly known as COVID-19. Till now there are approximately more than 10 million of people have suffered from this disease and lost their lives. The literature proved that there is no proper vaccine and drugs for the treatment of virus. Various in vitro and in silico studies should be considered and given a due attention to eradicate COVID-19. The in vitro data in most cases has been proved to be beneficial but more drugs and vaccine candidates should be studied and taken under consideration for future pandemic. Preventive measures need to be strictly adopted by the population across the world to protect themselves from the current pandemic. Combination of drugs and various inhibitors for blocking the entry and replication process should be considered. Simultaneously, identification of new treatment modalities needs to be investigated. Proper sharing of resources and knowledgeable information could prove to be helpful in fighting this deadly coronavirus disease. Present review will definitely lead us to understand the complexities of this disease and to design novel and potent drugs along with inhibitors against SARS-CoV-2. In addition, mechanistic insight may help us to understand virology of mutated strains of South Africa 501.V2 (variant), and UK variant.
References
Aanouz I, Belhassan A, El-Khatabi K, Lakhlifi T, El-Ldrissi M, Bouachrine M (2020) Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J Biomol Struct Dyn 1–9
Abbate A, Toldo S, Marchetti C, Kron J, Van Tassell BW, Dinarello CA (2020) Interleukin-1 and the inflammasome as therapeutic targets in cardiovascular disease. Circ Res 126(9):1260–1280
Abdul-Rahman A, Abdallah A, Yousif A, Memon SF (2020) Important considerations regarding the future management of Coronavirus (COVID-19). Int J Surg 79:6–7
Abruzzese E, Luciano L, D’Agostino F, Trawinska MM, Pane F, De Fabritiis P (2020) SARS-CoV-2 (COVID-19) and chronic myeloid leukemia (CML): a case report and review of ABL kinase involvement in viral infection. Mediterr J Hematol Infect Dis 12(1):e2020031
Akram M, Tahir IM, Shah SM, Mahmood Z, Altaf A, Ahmad K, Munir N, Daniyal M, Nasir S, Mehboob H (2018) Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: a systematic review. Phytother Res 32(5):811–822
Alas S, Emmanouilides C, Bonavida B (2001) Inhibition of interleukin 10 by rituximab results in down-regulation of bcl-2 and sensitization of B-cell non-Hodgkin’s lymphoma to apoptosis. Clin Canc Res 7(3):709–723
Alexander S, Armstrong J, Davenport A, Davies J, Faccenda E, Harding S, Levi-Schaffer F, Maguire J, Pawson A, Southan C, Spedding M (2020a) A rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development. Br J Pharmacol 177(21):4942–4966
Alexander PE, Debono VB, Mammen MJ, Iorio A, Aryal K, Deng D, Brocard E, Alhazzani W (2020b) COVID-19 coronavirus research has overall low methodological quality thus far: case in point for chloroquine/hydroxychloroquine. J Clin Epidemiol 123:120–126
Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP (2009) The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30(4):556–565
Al-Obaidi A, Elmezayen AD, Yelekçi K (2020) Homology modeling of human GABA-AT and devise some novel and potent inhibitors via computer-aided drug design techniques. J Biomol Struct Dyn 1–11
Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R (2003) Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300(5626):1763–1767
Arya R, Das A, Prashar V, Kumar M (2020) Potential inhibitors against papain-like protease of novel coronavirus (SARS-CoV-2) from FDA approved drugs. ChemRxiv Preprint. https://doi.org/10.26434/chemrxiv.11860011.v2
Astuti I, Ysrafil (2020) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr 14(4):407–412
Atal S, Fatima Z (2020) IL-6 inhibitors in the treatment of serious COVID-19: a promising therapy? Pharm Med 34:223–231
Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC (2005) The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79(24):15189–15198
Bartee E, McFadden G (2013) Cytokine synergy: an underappreciated contributor to innate anti-viral immunity. Cytokine 63(3):237–240
Benmerah A, Bayrou M, Cerf-Bensussan N, Dautry-Varsat A (1999) Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J Cell Sci 112(9):1303–1311
Berlin DA, Gulick RM, Martinez FJ (2020) Severe COVID-19. N Engl J Med. https://doi.org/10.1056/NEJMcp2009575
Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, Lucas JM, Nelson PS, Pöhlmann S, Soilleux EJ (2012) Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One 7(4):e35876
Bhardwaj VK, Singh R, Sharma J, Rajendran V, Purohit R, Kumar S (2020) Identification of bioactive molecules from Tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn 1–10
Bhatia R, Narang RK, Rawal RK (2020) Drug repurposing-a promising tool in drug discovery against CoV-19. Biomed J Sci Tech Res 28(5):21913–21915
Borkotoky S, Banerjee M (2020) A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem). J Biomol Struct Dyn 1–7
Burkard C, Verheije MH, Haagmans BL, van Kuppeveld FJ, Rottier PJ, Bosch BJ, de Haan CA (2015) ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J Virol 89(8):4434–4448
Burrell LM, Risvanis J, Kubota E, Dean RG, MacDonald PS, Lu S, Tikellis C, Grant SL, Lew RA, Smith AI, Cooper ME (2005) Myocardial infarction increases ACE2 expression in rat and humans. Eur Heart J 26(4):369–375
Carbajo-Lozoya J, Ma-Lauer Y, Malešević M, Theuerkorn M, Kahlert V, Prell E, Von Brunn B, Muth D, Baumert TF, Drosten C, Fischer G (2014) Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res 184:44–53
Chandel V, Raj S, Rathi B, Kumar D (2020a) In silico identification of potent FDA approved drugs against coronavirus COVID-19 main protease: a drug repurposing approach. Chem Biol Lett 7:166–175
Chandel V, Sharma PP, Raj S, Choudhari R, Rathi B, Kumar D (2020b) Structure-based drug repurposing for targeting Nsp9 replicase and spike proteins of severe acute respiratory syndrome coronavirus 2. J Biomol Struct Dyn 1–14.
Chang CK, Lo SC, Wang YS, Hou MH (2016) Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov Today 21(4):562–572
Chang L, Yan Y, Wang L (2020a) Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev 34(2):75–80
Chang YC, Tung YA, Lee KH, Chen TF, Hsiao YC, Chang HC, Hsieh TT, Su CH, Wang SS, Yu JY, Shih SS (2020b) Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking. Preprints 2020020242. https://doi.org/10.20944/preprints202002.0242.v1
Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z (2014) SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein cell 5(5):369–381
Chen IY, Moriyama M, Chang MF, Ichinohe T (2019) Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol 10:50. https://doi.org/10.3389/fmicb.2019.00050
Chen L, Xiong J, Bao L, Shi Y (2020) Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis 20:398–400
Cheng H, Wang Y, Wang GQ (2020) Organ-protective effect of angiotensin-converting enzyme 2 and its effect on the prognosis of COVID-19. J Med Virol 92(7):726–730
Chou CY, Chien CH, Han YS, Prebanda MT, Hsieh HP, Turk B, Chang GG, Chen X (2008) Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem Pharmacol 75(8):1601–1609
Choudhury S, Moulick D, Saikia P, Mazumder MK (2020) Evaluating the potential of different inhibitors on RNA-dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2: a molecular modeling approach. Med J Armed Forces India. https://doi.org/10.1016/j.mjafi.2020.05.005
Chu CM, Cheng VC, Hung IF, Wong MM, Chan KH, Chan KS, Kao RY, Poon LL, Wong CL, Guan Y, Peiris JS (2004) Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 59(3):252–256
Clinical Trials (2020) Evaluation of the efficacy and safety of Sarilumab in hospitalized patients with COVID-19. https://clinicaltrials.gov/ct2/show/NCT04315298
Clinicaltrial.gov (2020a) Favipiravir combined with tocilizumab in the treatment of corona virus disease 2019. Available at https://clinicaltrials.gov/ct2/show/NCT04310228
Clinicaltrial.gov (2020b) A study to evaluate the safety and efficacy of tocilizumab in patients with severe COVID‐19 pneumonia (COVACTA). Available at https://clinicaltrials.gov/ct2/show/NCT04320615
Clinicaltrial.gov (2020c) Tocilizumab in COVID‐19 pneumonia (TOCIVID‐19). Available at https://clinicaltrials.gov/ct2/show/NCT04317092
Clinicaltrials. Gov (2020) Treatments for covid-19: canadian arm of the solidarity trial (CATCO). Available from https://clinicaltrials.gov/ct2/show/NCT04330690
Cour M, Ovize M, Argaud L (2020) Cyclosporine A: a valid candidate to treat COVID-19 patients with acute respiratory failure? Crit Care 24(1):276
Danser AJ, Epstein M, Batlle D (2020) Renin-angiotensin system blockers and the COVID-19 pandemic: at present there is no evidence to abandon renin-angiotensin system blockers. Hypertension 75(6):1382–1385
De Clercq E (2002) Strategies in the design of antiviral drugs. Nat Rev Drug Discov 1(1):13–25
De Wilde AH, Jochmans D, Posthuma CC, Zevenhoven-Dobbe JC, Van Nieuwkoop S, Bestebroer TM, Van Den Hoogen BG, Neyts J, Snijder EJ (2014) Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother 58(8):4875–4884
Delang L, Abdelnabi R, Neyts J (2018) Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res 153:85–94
Dholaria BR, Bachmeier CA, Locke F (2019) Mechanisms and management of chimeric antigen receptor T-cell therapy-related toxicities. BioDrugs 33(1):45–60
Di Gennaro F, Pizzol D, Marotta C, Antunes M, Racalbuto V, Veronese N, Smith L (2020) Coronavirus diseases (COVID-19) current status and future perspectives: a narrative review. Int J Environ Res Public Health 17(8):2690
Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE (2009) A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin. Circ Res 87(5):1–9
Down B, Kulkarni S, Khan AH, Barker B, Tang I (2020) Novel coronavirus (COVID-19) infection: what a doctor on the frontline needs to know. Ann Med Surg (Lond) 55:24–29
Dutta D, Donaldson JG (2012) Search for inhibitors of endocytosis: intended specificity and unintended consequences. Cell Logist 2(4):203–208
Eifart P, Ludwig K, Böttcher C, de Haan CA, Rottier PJ, Korte T, Herrmann A (2007) The role of endocytosis and low pH in cell entry of the murine hepatitis virus MHV-A59. J Virol 81(19):10758–10768
Fang L, Karakiulakis G, Roth M (2020) Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med 8(4):e21
Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1282:1–23
Ferrario CM, Varagic J (2010) The ANG-(1–7)/ACE2/mas axis in the regulation of nephron function. Am J Physiol Renal Physiol 298(6):F1297–F1305
Franchini M, Marano G, Velati C, Pati I, Pupella S, Liumbruno GM (2020) Operational protocol for donation of anti‐COVID‐19 convalescent plasma in Italy. Vox Sang 12940. https://doi.org/10.1111/vox.12940
Fujimoto LM, Roth R, Heuser JE, Schmid SL (2020) Actin assembly plays a variable, but not obligatory role in receptor-mediated endocytosis. Traffic 1(2):161–171
Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, Morrey JD (2009) T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res 82(3):95–102
Furuta Y, Komeno T, Nakamura T (2017) Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci 93(7):449–463
Gabellini C, Trisciuoglio D, Desideri M, Candiloro A, Ragazzoni Y, Orlandi A, Zupi G, Del Bufalo D (2009) Functional activity of CXCL8 receptors, CXCR1 and CXCR2, on human malignant melanoma progression. Eur J Cancer 45(14):2618–2627
Gautret P, Lagier JC, Parola P, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, Dupont HT, Honoré S et al (2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 56(1):105949
Ghosh AK, Takayama J, Rao KV, Ratia K, Chaudhuri R, Mulhearn DC, Lee H, Nichols DB, Baliji S, Baker SC, Johnson ME, Mesecar AD (2010) Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein−ligand X-ray structure and biological evaluation. J Med Chem 53(13):4968–4979
Gibson AE, Noel RJ, Herlihy JT, Ward WF (1989) Phenylarsine oxide inhibition of endocytosis: effects on asialofetuin internalization. Am J Physiol 257:C182-184
Gil Ayuso-Gontán C, Ginex T, Maestro I, Nozal V, Barrado-Gil L, Cuesta-Geijo MÁ, Urquiza J, Ramírez D, Alonso C, Campillo NE, Martinez A (2020) COVID-19: drug targets and potential treatments. J Med Chem 63(21):12359–12386
Greene SJ, Gheorghiade M, Vaduganathan M, Ambrosy AP, Mentz RJ, Subacius H, Maggioni AP, Nodari S, Konstam MA, Butler J, Filippatos G (2013) Haemoconcentration, renal function, and post-discharge outcomes among patients hospitalized for heart failure with reduced ejection fraction: insights from the EVEREST trial. Eur J Heart Fail 15(12):1401–1411
Grein J, Ohmagari N, Shin D et al (2020) Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med 382:2327–2336
Gunn BM, Yu WH, Karim MM, Brannan JM, Herbert AS, Wec AZ, Halfmann PJ, Fusco ML, Schendel SL, Gangavarapu K, Krause T (2018) A role for Fc function in therapeutic monoclonal antibody-mediated protection against Ebola virus. Cell Host Microbe 24(2):221–233
Guo Y, Korteweg C, McNutt MA, Gu J (2008) Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res 133(1):4–12
Haber PK, Ye M, Wysocki J, Maier C, Haque SK, Batlle D (2014) Angiotensin-converting enzyme 2-independent action of presumed angiotensin-converting enzyme 2 activators: studies in vivo, ex vivo and in vitro. Hypertension 63(4):774–782
Hage R, Steinack C, Schuurmans M (2020) Calcineurin inhibitors in COVID-19-lessons learnt from transplantation medicine. Biomed J Sci Tech Res 29(3):22447–22448
Hamming I, Timens W, Bulthuis ML, Lely AT, Navis GV, van Goor H (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203(2):631–637
Han YS, Chang GG, Juo CG, Lee HJ, Yeh SH, Hsu JT, Chen X (2005) Papain-like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): expression, purification, characterization and inhibition. Biochemistry 44(30):10349–10359
Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S (2014) TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol 88(2):1293–1307
Hilgenfeld R (2014) From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J 281(18):4085–4096
Hirota M, Shimosegawa T, Kitamura K, Takeda K, Takeyama Y, Mayumi T, Ito T, Takenaka M, Iwasaki E, Sawano H, Ishida E et al (2020) Continuous regional arterial infusion versus intravenous administration of the protease inhibitor nafamostat mesilate for predicted severe acute pancreatitis: a multicenter, randomized, open-label, phase 2 trial. J Gastroenterol 55(3):342–352
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271-280.e8
Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G (2020) First case of 2019 novel coronavirus in the United States. N Eng J Med 382:929–936. https://doi.org/10.1056/NEJMoa2001191
Hosseini-Zare MS, Thilagavathi R, Selvam C (2020) Targeting severe acute respiratory syndrome-coronavirus (SARS-CoV-1) with structurally diverse inhibitors: a comprehensive review. RSC Adv 10(47):28287–28299
Hu X, Li WP, Meng C, Ivashkiv LB (2003) Inhibition of IFN-γ signaling by glucocorticoids. J Immunol 170(9):4833–4839
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z (2019) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506
Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, Crackower MA (2005) Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436(7047):112–116
Inoue Y, Tanaka N, Tanaka Y, Inoue S, Morita K, Zhuang M, Hattori T, Sugamura K (2007) Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol 81(16):8722–8729
Ishiyama Y, Gallagher PE, Averill DB, Tallant EA, Brosnihan KB, Ferrario CM (2004) Upregulation of angiotensin-converting enzyme 2 after myocardial infarction by blockade of angiotensin II receptors. Hypertension 43(5):970–976
Jeon S, Ko M, Lee J, Choi I, Byun SY et al (2020) Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother 64(7):e00819–e00820
Jiang F, Deng L, Zhang L, Cai Y, Cheung CW, Xia Z (2020) Review of the clinical characteristics of coronavirus disease 2019 (COVID-19). J Gen Intern Med 35:1545–1549
Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J (2013) The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS One 8(7):e68347
Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP, Fang C, Huang D, Huang LQ, Huang Q, Han Y et al (2020) A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil Med Res 7:4. https://doi.org/10.1186/s40779-020-0233-6
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J (2016) PubChem substance and compound databases. Nucleic Acids Res 44(D1):D1202–D1213
Kruse RL (2020) Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res 9:72
Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L et al (2005) A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 11(8):875–879
Kumar A, Choudhir G, Shukla SK, Sharma M, Tyagi P, Bhushan A, Rathore M (2020) Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J Biomol Struct Dyn 1–11
Kupferschmidt K, Cohen J (2020) Race to find COVID-19 treatments accelerates. Science 367(6485):1412–1413
Lagana A, Vadnais J, Le PU, Nguyen TN, Laprade R, Nabi IR, Noel J (2000) Regulation of the formation of tumor cell pseudopodia by the Na (+)/H (+) exchanger NHE1. J Cell Sci 113:3649–3662
Larkin JM, Brown MS, Goldstein JL, Anderson RG (1983) Depletion of intracellular potassium arrests coated pit formation and receptor-mediated endocytosis in fibroblasts. Cell 33(1):273–285
Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Ménard R (2005) The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79(24):15199–15208
Liu B, Zhou J (2005) SARS-CoV protease inhibitors design using virtual screening method from natural products libraries. J Comput Chem 26(5):484–490
Liu SH, Marks MS, Brodsky FM (1998) A dominant-negative clathrin mutant differentially affects trafficking of molecules with distinct sorting motifs in the class II major histocompatibility complex (MHC) pathway. J Cell Biol 140(5):1023–1037
Liu K, Fang YY, Deng Y, Liu W, Wang MF, Ma JP, Xiao W, Wang YN, Zhong MH, Li CH, Li GC (2020) Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl) 133(9):1025–1031
Lloyd CM, Hessel EM (2010) Functions of T cells in asthma: more than just TH 2 cells. Nat Rev Immunol 10(12):838–848
Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T (2006) Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10(6):839–850
Maurya VK, Kumar S, Bhatt ML, Saxena SK (2020) Therapeutic development and drugs for the treatment of COVID-19. In: Coronavirus disease 2019 (COVID-19), pp 109–126. https://doi.org/10.1007/978-981-15-4814-7_10
McBride R, Van Zyl M, Fielding BC (2014) The coronavirus nucleocapsid is a multifunctional protein. Viruses 6(8):2991–3018
Mistry P, Reid J, Pouliquen I, McHugh S, Abberley L, DeWall S, Taylor A, Tong X, Rocha MD, McKie E (2014) Safety, tolerability, pharmacokinetics, and pharmacodynamics of single-dose antiinterleukin-18 mAb GSK1070806 in healthy and obese subjects. Int J Clin Pharmacol Ther 52(10):867–879
Mulangu S, Dodd LE, Davey RT Jr, Tshiani Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D, Tshomba Oloma A, Ibanda A, Ali R (2019) A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med 381(24):2293–2303
Mullard A (2020) COVID-19 vaccine development pipeline gears up. World Rep 395:1751–1752
Mummed Y (2020) Molecular targets for COVID-19 drug development: enlightening Nigerians about the pandemic and future treatment. Biosaf Health. https://doi.org/10.1016/j.bsheal.2020.07.002
Muramatsu T, Takemoto C, Kim YT, Wang H, Nishii W, Terada T, Shirouzu M, Yokoyama S (2016) SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc Natl Acad Sci USA 113(46):12997–13002
Ocaranza MP, Godoy I, Jalil JE, Varas M, Collantes P, Pinto M, Roman M, Ramirez C, Copaja M, Diaz-Araya G, Castro P (2006) Enalapril attenuates downregulation of angiotensin-converting enzyme 2 in the late phase of ventricular dysfunction in myocardial infarcted rat. Hypertension 48(4):572–578
O’Connor KA, Roth BL (2005) Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat Rev Drug Discov 4(12):1005–1014
Oh CK, Leigh R, McLaurin KK, Kim K, Hultquist M, Molfino NA (2013) A randomized, controlled trial to evaluate the effect of an anti-interleukin-9 monoclonal antibody in adults with uncontrolled asthma. Respir Res 14(1):93. https://doi.org/10.1186/1465-9921-14-93
Ohtsuki M, Morimoto SI, Izawa H, Ismail TF, Ishibashi-Ueda H, Kato Y, Horii T, Isomura T, Suma H, Nomura M, Hishida H (2010) Angiotensin converting enzyme 2 gene expression increased compensatory for left ventricular remodeling in patients with end-stage heart failure. Int J Cardio 145:333–334
Omrani AS, Saad MM, Baig K, Bahloul A, Abdul-Matin M, Alaidaroos AY, Almakhlafi GA, Albarrak MM, Memish ZA, Albarrak AM (2014) Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis 14(11):1090–1095
Perico L, Benigni A, Remuzzi G (2020) Should COVID-19 concern nephrologists? Why and to what extent? The emerging impasse of angiotensin blockade. Nephron 144(5):213–221
Prajapat M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, Kumar S, Bhattacharyya A, Kumar H, Bansal S, Medhi B (2020) Drug targets for corona virus: a systematic review. Indian J Pharmacol 52(1):56–65
Qiu Z, Hingley ST, Simmons G, Yu C, Sarma JD, Bates P, Weiss SR (2006) Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J Virol 80(12):5768–5776
Raj S, Chandel V, Rathi B, Kumar D (2020) Understanding the Molecular Mechanism(s) of SARS-CoV2 Infection and propagation in human to discover potential preventive and therapeutic approach. Coronaviruses. 1:73–81. https://doi.org/10.20944/preprints202004.0285.v1
Rana S, Sharma S, Ghosh KS (2020) Virtual screening of naturally occurring antiviral molecules for SARS-CoV-2 mitigation using docking tool on multiple molecular targets. ChemRxiv Preprint. https://doi.org/https://doi.org/10.26434/chemrxiv.12403940.v1
Rossi GP, Sanga V, Barton M (2020) Potential harmful effects of discontinuing ACE-inhibitors and ARBs in COVID-19 patients. Elife 9:e57278
Rudnicka L, Goldust M, Glowacka P, Sikora M, Sar-Pomian M, Rakowska A, Samochocki Z, Olszewska M (2020) Cyclosporine therapy during the COVID-19 pandemic is not a reason for concern. J Am Acad Dermatol 83(2):e151–e152
Sai JK, Suyama M, Kubokawa Y, Matsumura Y, Inami K, Watanabe S (2010) Efficacy of camostat mesilate against dyspepsia associated with non-alcoholic mild pancreatic disease. J Gastroenterol 45(3):335–341
Saif LJ (2004) Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev Sci Tech 23:643–660
Saikatendu KS, Joseph JS, Subramanian V, Clayton T, Griffith M, Moy K, Velasquez J, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P (2005) Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13(11):1665–1675
Sanchez-Pernaute O, Romero-Bueno FI, Selva-O’Callaghan A (2020) Why choose cyclosporin A as first-line therapy in COVID-19 pneumonia. Reumatol Clin. https://doi.org/10.1016/j.reuma.2020.03.001
Santos RA, Sampaio WO, Alzamora AC, Motta-Santos D, Alenina N, Bader M, Campagnole-Santos MJ (2018) The ACE2/angiotensin-(1–7)/MAS axis of the renin-angiotensin system: focus on angiotensin-(1–7). Physiol Rev 98(1):505–553
Saxena A (2020) Drug targets for COVID-19 therapeutics: ongoing global efforts. J Biosci 45(1):87
Schlegel R, Dickson RB, Willingham MC, Pastan IH (1982) Amantadine and dansylcadaverine inhibit vesicular stomatitis virus uptake and receptor-mediated endocytosis of alpha 2-macroglobulin. Proc Natl Acad Sci USA 79:2291–2295
Scott LJ (2017) Tocilizumab: a review in rheumatoid arthritis. Drugs 77:1865–1879
Shakoory B, Carcillo JA, Chatham WW, Amdur RL, Zhao H, Dinarello CA, Cron RQ, Opal SM (2016) Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of the macrophage activation syndrome: re-analysis of a prior phase III trial. Crit Care Med 44(2):275
Siegel D, Hui HC, Doerffler E, Clarke MO, Chun K, Zhang L, Neville S, Carra E, Lew W, Ross B, Wang Q (2017) Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo [2, 1-f][triazin-4-amino] adenine c-nucleoside (gs-5734) for the treatment of Ebola and emerging viruses. J Med Chem 60(5):1648–1661
Singh S, Florez H (2020) Coronavirus disease 2019 drug discovery through molecular docking. F1000Res 9:502
Sissoko D, Laouenan C, Folkesson E, Mlebing AB, Beavogui AH, Baize S, Camara AM, Maes P, Shepherd S, Danel C, Carazo S (2016) Experimental treatment with favipiravir for Ebola virus disease (The JIKI Trial): a historically controlled, single-arm proof-of concept trial in Guinea. PLOS Med 13(6):e1002066
Siu KL, Yuen KS, Castano-Rodriguez C, Ye ZW, Yeung ML, Fung SY, Yuan S, Chan CP, Yuen KY, Enjuanes L, Jin DY (2019) Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J 33(8):8865–8877
Sood S, Aggarwal V, Aggarwal D, Upadhyay SK, Sak K, Tuli HS, Kumar M, Kumar J, Talwar S (2020) COVID-19 pandemic: from molecular biology, pathogenesis, detection and treatment to global societal impact. Curr Pharmacol Rep 6:212–227
Soro-Paavonen A, Gordin D, Forsblom C, Rosengard-Barlund M, Waden J, Thorn L, Sandholm N, Thomas MC, Groop PH, FinnDiane Study Group (2012) Circulating ACE2 activity is increased in patients with type 1 diabetes and vascular complications. J Hypertens 30(2):375–383
Stebbing J, Phelan A, Griffin I, Tucker C, Oechsle O, Smith D, Richardson P (2020) COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis 20(4):400–402
Sydnes MO, Hayashi Y, Sharma VK, Hamada T, Bacha U, Barrila J, Freire E, Kiso Y (2006) Synthesis of glutamic acid and glutamine peptides possessing a trifluoromethyl ketone group as SARS-CoV 3CL protease inhibitors. Tetrahedron 62(36):8601–8609
Talwar S, Sood S, Kumar J, Chauhan R, Sharma M, Tuli HS (2020) Ayurveda and allopathic therapeutic strategies in coronavirus pandemic treatment 2020. Curr Pharmacol Rep 22:1–10
Tanaka Y, Sato Y, Sasaki T (2013) Suppression of coronavirus replication by cyclophilin inhibitors. Viruses 5:1250–1260
Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LF (2020) The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 20(6):363–374
Thomas PG, Dash P, Aldridge JR Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, Doherty PC (2009) The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30(4):566–575
Tuley A, Fast W (2018) The taxonomy of covalent inhibitors. Biochemistry 57(24):3326–3337
Turk B (2006) Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov 5(9):785–799
Ucciferri C, Auricchio A, Di Nicola M, Potere N, Abbate A, Cipollone F, Vecchiet J, Falasca K (2020) Canakinumab in a subgroup of patients with COVID-19. Lancet Rheumatol 2(8):e457–e458
ul Qamar MT, Alqahtani SM, Alamri MA, Chen LL (2020) Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal 10(4):313–319
Upadhyay SK, Singh R, Singh M, Kumar V, Yadav M, Aggarwal D, Sehrawat N (2020) COVID-19 in republic of India: A report on situation and precautionary strategies to global pandemic. Bull Environ Pharmacol Life Sci 9:39–48
Vaduganathan M, Vardeny O, Michel T, McMurray JJ, Pfeffer MA, Solomon SD (2020) Renin–angiotensin–aldosterone system inhibitors in patients with Covid-19. N Engl J Med 382(17):1653–1659
Van Der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL (1993) Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol 122(3):553–563
Van Erp EA, Luytjes W, Ferwerda G, Van Kasteren PB (2019) Fc-mediated antibody effector functions during respiratory syncytial virus infection and disease. Front Immunol 10:548
Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J, Godbout K, Parsons T, Baronas E, Hsieh F, Acton S (2002) Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem 277(17):14838–14843
Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, Ksiazek TG (2005) Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J 2:69
Von Kleist L, Stahlschmidt W, Bulut H, Gromova K, Puchkov D, Robertson MJ, MacGregor KA, Tomilin N, Pechstein A, Chau N, Chircop M (2011) Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 146(3):471–484
Wahedi HM, Ahmad S, Abbasi SW (2020) Stilbene-based natural compounds as promising drug candidates against COVID-19. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1762743
Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30(3):269–271
Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N (2016) Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531(7594):381–385
Willicombe M, Thomas D, McAdoo S (2020) COVID-19 and calcineurin inhibitors: should they get left out in the storm? J Am Soc Nephrol 31(6):1145–1146
Wösten-van Asperen RM, Lutter R, Specht PA, Moll GN, van Woensel JB, van der Loos CM, van Goor H, Kamilic J, Florquin S, Bos AP (2011) Acute respiratory distress syndrome leads to reduced ratio of ACE/ACE2 activities and is prevented by angiotensin-(1–7) or an angiotensin II receptor antagonist. J Pathol 225(4):618–627
Xu X, Han M, Li T, Sun W, Wang D, Fu B, Zhou Y, Zheng X, Yang Y, Li X, Zhang X (2020a) Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA 117(20):10970–10975
Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y (2020b) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8:420–422
Xudong X, Junzhu C, Xingxiang W, Furong Z, Yanrong L (2006) Age-and gender-related difference of ACE2 expression in rat lung. Life Sci 78(19):2166–2171
Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, Li J, Zhou Z, Ding Y, Zhao Q, Zhang XC (2008) Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J virol 82(5):2515–2527
Yamamoto M, Matsuyama S, Li X, Takeda M, Kawaguchi Y, Inoue JI, Matsuda Z (2016) Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome coronavirus S protein mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother 60(11):6532–6539
Yamaya M, Shimotai Y, Hatachi Y, Kalonji NL, Tando Y, Kitajima Y, Matsuo K, Kubo H, Nagatomi R, Hongo S, Homma M (2015) The serine protease inhibitor camostat inhibits influenza virus replication and cytokine production in primary cultures of human tracheal epithelial cells. Pulm Pharmacol Ther 33:66–74
Yang N, Shen HM (2020) Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int J Biol Sci 16:1724–1731
Yang H, Bartlam M, Rao Z (2006) Drug design targeting the main protease, the Achilles’ heel of coronaviruses. Curr Pharm Des 12:4573–4590
Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS (2020) Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intens Care Med 46(4):586–590
Zhao X, Greener T, Al-Hasani H, Cushman SW, Eisenberg E, Greene LE (2001) Expression of auxilin or AP180 inhibits endocytosis by mislocalizingclathrin: evidence for formation of nascent pits containing AP1 or AP2 but not clathrin. J Cell Sci 114:353–365
Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R Jr, Nunneley JW, Barnard D, Pöhlmann S, McKerrow JH, Renslo AR, Simmons G (2015) Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res 116:76–84
Zhu L, George S, Schmidt MF, Al-Gharabli SI, Rademann J, Hilgenfeld R (2011) Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res 92(2):204–212
Ziebuhr J (2005) The coronavirus replicase. Curr Top Microbiol Immunol 287:57–94
Ziebuhr J, Snijder EJ, Gorbalenya AE (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J General Virol 81(4):853–879
Acknowledgements
The authors are grateful to the Maharishi Markandeshwar (Deemed to be University) for providing the requisite platform for this work.
Author information
Authors and Affiliations
Contributions
HST designed and collected the data; SS contributed in abstract, introduction, and conclusion; JK, PK, PS, SP, and PY contributed in mechanisms of action of drugs; AKS, DA, and KS proof read the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Rights and permissions
About this article
Cite this article
Tuli, H.S., Sood, S., Kaur, J. et al. Mechanistic insight into anti-COVID-19 drugs: recent trends and advancements. 3 Biotech 11, 110 (2021). https://doi.org/10.1007/s13205-021-02644-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02644-8