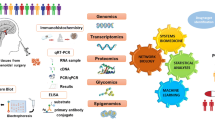
Abstract
Nonfuctional pituitary neuroendocrine tumor (NF-PitNET) is highly heterogeneous and generally considered a common intracranial tumor. A series of molecules are involved in NF-PitNET pathogenesis that alter in multiple levels of genome, transcriptome, proteome, and metabolome, and those molecules mutually interact to form dynamically associated molecular-network systems. This article reviewed signaling pathway alterations in NF-PitNET based on the analyses of the genome, transcriptome, proteome, and metabolome, and emphasized signaling pathway network alterations based on the integrative omics, including calcium signaling pathway, cGMP-PKG signaling pathway, mTOR signaling pathway, PI3K/AKT signaling pathway, MAPK (mitogen-activated protein kinase) signaling pathway, oxidative stress response, mitochondrial dysfunction, and cell cycle dysregulation, and those signaling pathway networks are important for NF-PitNET formation and progression. Especially, this review article emphasized the altered signaling pathways and their key molecules related to NF-PitNET invasiveness and aggressiveness that are challenging clinical problems. Furthermore, the currently used medication and potential therapeutic agents that target these important signaling pathway networks are also summarized. These signaling pathway network changes offer important resources for insights into molecular mechanisms, discovery of effective biomarkers, and therapeutic targets for patient stratification, predictive diagnosis, prognostic assessment, and targeted therapy of NF-PitNET.





Similar content being viewed by others
Data Availability
All data and materials are available in the current manuscript.
Code availability
Not applicable.
References
Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS Statistical Report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–62. https://doi.org/10.1093/neuonc/nov189.
Molitch ME. Diagnosis and treatment of pituitary adenomas: a review. JAMA. 2017;317:516–24. https://doi.org/10.1001/jama.2016.19699.
Andela CD, Lobatto DJ, Pereira AM, van Furth WR, Biermasz NR. How non-functioning pituitary adenomas can affect health-related quality of life: a conceptual model and literature review. Pituitary. 2018;21:208–16. https://doi.org/10.1007/s11102-017-0860-4.
Greenman Y, Stern N. Non-functioning pituitary adenomas. Best Pract Res Clin Endocrinol Metab. 2009;23:625–38. https://doi.org/10.1016/j.beem.2009.05.005.
Tampourlou M, Fountas A, Ntali G, Karavitaki N. Mortality in patients with non-functioning pituitary adenoma. Pituitary. 2018;21(2):203–7. https://doi.org/10.1007/s11102-018-0863-9.
Mete O, Lopes MB. Overview of the 2017 WHO classification of pituitary tumors. Endocr Pathol. 2017;28:228–43. https://doi.org/10.1007/s12022-017-9498-z.
Drummond J, Roncaroli F, Grossman AB, Korbonits M. Clinical and pathological aspects of silent pituitary adenomas. J Clin Endocrinol Metab. 2019;104:2473–89. https://doi.org/10.1210/jc.2018-00688.
Scheithauer BW, Kovacs KT, Laws ER Jr, Randall RV. Pathology of invasive pituitary tumors with special reference to functional classification. J Neurosurg. 1986;65:733–44. https://doi.org/10.3171/jns.1986.65.6.0733.
Wang H, Chen K, Yang Z, Li W, Wang C, Zhang G, et al. Diagnosis of invasive nonfunctional pituitary adenomas by serum extracellular vesicles. Anal Chem. 2019;91:9580–9. https://doi.org/10.1021/acs.analchem.9b00914.
Long Y, Lu M, Cheng T, Zhan X, Zhan X. Multiomics-based signaling pathway network alterations in human non-functional pituitary adenomas. Front Endocrinol (Lausanne). 2019;10:835. https://doi.org/10.3389/fendo.2019.00835.
Wu B, Jiang S, Wang X, Zhong S, Bi Y, Yi D, Liu G, Hu D, Dou G, Chen Y, Wu Y, Dong J. Identification of driver genes and key pathways of non-functional pituitary adenomas predicts the therapeutic effect of STO-609. PLoS One. 2020;15:e0240230. https://doi.org/10.1371/journal.pone.0240230.
Joshi H, Vastrad B, Vastrad C. Identification of important invasion-related genes in non-functional pituitary adenomas. J Mol Neurosci. 2019;68:565–89. https://doi.org/10.1007/s12031-019-01318-8.
Song W, Qian L, Jing G, Jie F, Xiaosong S, Chunhui L, et al. Aberrant expression of the sFRP and WIF1 genes in invasive non-functioning pituitary adenomas. Mol Cell Endocrinol. 2018;474:168–75. https://doi.org/10.1016/j.mce.2018.03.005.
Esapa CT, Harris PE. Mutation analysis of protein kinase A catalytic subunit in thyroid adenomas and pituitary tumours. Eur J Endocrinol. 1999;141:409–12. https://doi.org/10.1530/eje.0.1410409.
Farrell WE, Talbot JA, Bicknell EJ, Simpson D, Clayton RN. Genomic sequence analysis of a key residue (Arg183) in human G alpha q in invasive non-functional pituitary adenomas. Clin Endocrinol (Oxf). 1997;47:241–4. https://doi.org/10.1046/j.1365-2265.1997.2891088.x.
Faccenda E, Melmed S, Bevan JS, Eidne KA. Structure of the thyrotrophin-releasing hormone receptor in human pituitary adenomas. Clin Endocrinol (Oxf). 1996;44:341–7. https://doi.org/10.1046/j.1365-2265.1996.684506.x.
Zhu H, Guo J, Shen Y, Dong W, Gao H, Miao Y, et al. Functions and mechanisms of tumor necrosis factor-α and noncoding RNAs in bone-invasive pituitary adenomas. Clin Cancer Res. 2018;24:5757–66. https://doi.org/10.1158/1078-0432.ccr-18-0472.
Taniguchi-Ponciano K, Gomez-Apo E, Chavez-Macias L, Vargas G, Espinosa-Cardenas E, Ramirez-Renteria C, et al. Molecular alterations in non-functioning pituitary adenomas. Cancer Biomark. 2020;28:193–9. https://doi.org/10.3233/cbm-191121.
Kim YH, Kim JH. Transcriptome Analysis identifies an attenuated local immune response in invasive nonfunctioning pituitary adenomas. Endocrinol Metab (Seoul). 2019;34:314–22. https://doi.org/10.3803/EnM.2019.34.3.314.
Wu S, Gu Y, Huang Y, Wong TC, Ding H, Liu T, et al. Novel biomarkers for non-functioning invasive pituitary adenomas were identified by using analysis of microRNAs expression profile. Biochem Genet. 2017;55:253–67. https://doi.org/10.1007/s10528-017-9794-9.
Xing W, Qi Z. Genome-wide identification of lncRNAs and mRNAs differentially expressed in non-functioning pituitary adenoma and construction of an lncRNA-mRNA co-expression network. Biol Open. 2019;8(1):bio037127. https://doi.org/10.1242/bio.037127.
Guo J, Fang Q, Liu Y, Xie W, Zhang Y, Li C. Identifying critical protein-coding genes and long non-coding RNAs in non-functioning pituitary adenoma recurrence. Oncol Lett. 2021;21:264. https://doi.org/10.3892/ol.2021.12525.
Li J, Li C, Wang J, Song G, Zhao Z, Wang H, et al. Genome-wide analysis of differentially expressed lncRNAs and mRNAs in primary gonadotrophin adenomas by RNA-seq. Oncotarget. 2017;8:4585–606. https://doi.org/10.18632/oncotarget.13948.
Lee M, Marinoni I, Irmler M, Psaras T, Honegger JB, Beschorner R, et al. Transcriptome analysis of MENX-associated rat pituitary adenomas identifies novel molecular mechanisms involved in the pathogenesis of human pituitary gonadotroph adenomas. Acta Neuropathol. 2013;126:137–50. https://doi.org/10.1007/s00401-013-1132-7.
Richardson TE, Shen ZJ, Kanchwala M, Xing C, Filatenkov A, Shang P, et al. Aggressive behavior in silent subtype III pituitary adenomas may depend on suppression of local immune response: a whole transcriptome analysis. J Neuropathol Exp Neurol. 2017;76:874–82. https://doi.org/10.1093/jnen/nlx072.
Zhan X, Desiderio DM. Signaling pathway networks mined from human pituitary adenoma proteomics data. BMC Med Genomics. 2010;3:13. https://doi.org/10.1186/1755-8794-3-13.
Qian S, Zhan X, Lu M, Li N, Long Y, Li X, et al. Quantitative analysis of ubiquitinated proteins in human pituitary and pituitary adenoma tissues. Front Endocrinol (Lausanne). 2019;10:328. https://doi.org/10.3389/fendo.2019.00328.
Li J, Wen S, Li B, Li N, Zhan X. Phosphorylation-mediated molecular pathway changes in human pituitary neuroendocrine tumors identified by quantitative phosphoproteomics. Cells. 2021;10(9):2225. https://doi.org/10.3390/cells10092225.
Zhan X, Desiderio DM, Wang X, Zhan X, Guo T, Li M, et al. Identification of the proteomic variations of invasive relative to non-invasive non-functional pituitary adenomas. Electrophoresis. 2014;35:2184–94. https://doi.org/10.1002/elps.201300590.
Zhan X, Wang X, Long Y, Desiderio DM. Heterogeneity analysis of the proteomes in clinically nonfunctional pituitary adenomas. BMC Med Genomics. 2014;7:69. https://doi.org/10.1186/s12920-014-0069-6.
Wang X, Guo T, Peng F, Long Y, Mu Y, Yang H, et al. Proteomic and functional profiles of a follicle-stimulating hormone positive human nonfunctional pituitary adenoma. Electrophoresis. 2015;36:1289–304. https://doi.org/10.1002/elps.201500006.
Wang Y, Cheng T, Lu M, Mu Y, Li B, Li X, et al. TMT-based quantitative proteomics revealed follicle-stimulating hormone (FSH)-related molecular characterizations for potentially prognostic assessment and personalized treatment of FSH-positive non-functional pituitary adenomas. EPMA J. 2019;10:395–414. https://doi.org/10.1007/s13167-019-00187-w.
Simpson DJ, Frost SJ, Bicknell JE, Broome JC, McNicol AM, Clayton RN, et al. Aberrant expression of G(1)/S regulators is a frequent event in sporadic pituitary adenomas. Carcinogenesis. 2001;22:1149–54. https://doi.org/10.1093/carcin/22.8.1149.
Cheng S, Xie W, Miao Y, Guo J, Wang J, Li C, et al. Identification of key genes in invasive clinically non-functioning pituitary adenoma by integrating analysis of DNA methylation and mRNA expression profiles. J Transl Med. 2019;17(1):407. https://doi.org/10.1186/s12967-019-02148-3.
Wei Z, Zhou C, Li M, Huang R, Deng H, Shen S, et al. Integrated multi-omics profiling of nonfunctioning pituitary adenomas. Pituitary. 2021;24:312–25. https://doi.org/10.1007/s11102-020-01109-0.
Yu SY, Hong LC, Feng J, Wu YT, Zhang YZ. Integrative proteomics and transcriptomics identify novel invasive-related biomarkers of non-functioning pituitary adenomas. Tumour Biol. 2016;37:8923–30. https://doi.org/10.1007/s13277-015-4767-2.
Feng J, Yu SY, Li CZ, Li ZY, Zhang YZ. Integrative proteomics and transcriptomics revealed that activation of the IL-6R/JAK2/STAT3/MMP9 signaling pathway is correlated with invasion of pituitary null cell adenomas. Mol Cell Endocrinol. 2016;436:195–203. https://doi.org/10.1016/j.mce.2016.07.025.
Cheng T, Wang Y, Lu M, Zhan X, Zhou T, Li B, et al. Quantitative analysis of proteome in non-functional pituitary adenomas: clinical relevance and potential benefits for the patients. Front Endocrinol (Lausanne). 2019;10:854. https://doi.org/10.3389/fendo.2019.00854.
Liu D, Li J, Li N, Lu M, Wen S, Zhan X. Integration of quantitative phosphoproteomics and transcriptomics revealed phosphorylation-mediated molecular events as useful tools for a potential patient stratification and personalized treatment of human nonfunctional pituitary adenomas. EPMA J. 2020;11:419–67. https://doi.org/10.1007/s13167-020-00215-0.
Wen S, Li J, Yang J, Li B, Li N, Zhan X. Quantitative acetylomics revealed acetylation-mediated molecular pathway network changes in human nonfunctional pituitary neuroendocrine tumors. Front Endocrinol (Lausanne). 2021;12:753606. https://doi.org/10.3389/fendo.2021.753606.
Wang Z, Guo X, Wang W, Gao L, Bao X, Feng M, et al. UPLC-MS/MS-based lipidomic profiles revealed aberrant lipids associated with invasiveness of silent corticotroph adenoma. J Clin Endocrinol Metab. 2021;106:e273–87. https://doi.org/10.1210/clinem/dgaa708.
Feng J, Gao H, Zhang Q, Zhou Y, Li C, Zhao S, et al. Metabolic profiling reveals distinct metabolic alterations in different subtypes of pituitary adenomas and confers therapeutic targets. J Transl Med. 2019;17:291. https://doi.org/10.1186/s12967-019-2042-9.
Panda AC, Abdelmohsen K, Gorospe M. SASP regulation by noncoding RNA. Mech Ageing Dev. 2017;168:37–43. https://doi.org/10.1016/j.mad.2017.05.004.
Yang Q, Wang Y, Zhang S, Tang J, Li F, Yin J, et al. Biomarker discovery for immunotherapy of pituitary adenomas: enhanced robustness and prediction ability by modern computational tools. Int J Mol Sci. 2019;20(1):151. https://doi.org/10.3390/ijms20010151.
Monti C, Zilocchi M, Colugnat I, Alberio T. Proteomics turns functional. J Proteomics. 2019;198:36–44. https://doi.org/10.1016/j.jprot.2018.12.012.
Karimi P, Shahrokni A, Ranjbar MR. Implementation of proteomics for cancer research: past, present, and future. Asian Pac J Cancer Prev. 2014;15:2433–8. https://doi.org/10.7314/apjcp.2014.15.6.2433.
Li N, Desiderio DM, Zhan X. The use of mass spectrometry in a proteome-centered multiomics study of human pituitary adenomas. Mass Spectrom Rev. 2021. https://doi.org/10.1002/mas.21710.
Walsh MT, Couldwell WT. Symptomatic cystic degeneration of a clinically silent corticotroph tumor of the pituitary gland. Skull Base. 2010;20:367–70. https://doi.org/10.1055/s-0030-1253579.
Zhou W, Song Y, Xu H, Zhou K, Zhang W, Chen J, et al. In nonfunctional pituitary adenomas, estrogen receptors and slug contribute to development of invasiveness. J Clin Endocrinol Metab. 2011;96:E1237-1245. https://doi.org/10.1210/jc.2010-3040.
Hu R, Wang X, Zhan X. Multi-parameter systematic strategies for predictive, preventive and personalised medicine in cancer. EPMA J. 2013;4:2. https://doi.org/10.1186/1878-5085-4-2.
Zhan X, Desiderio DM. Comparative proteomics analysis of human pituitary adenomas: current status and future perspectives. Mass Spectrom Rev. 2005;24:783–813. https://doi.org/10.1002/mas.20039.
Rinschen MM, Ivanisevic J. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol. 2019;20:353–67. https://doi.org/10.1038/s41580-019-0108-4.
Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–9. https://doi.org/10.1038/nrm.2016.25.
Hu C, Zhou Y, Feng J, Zhou S, Li C, Zhao S, et al. Untargeted lipidomics reveals specific lipid abnormalities in nonfunctioning human pituitary adenomas. J Proteome Res. 2020;19:455–63. https://doi.org/10.1021/acs.jproteome.9b00637.
Ijare OB, Baskin DS, Pichumani K. Ex Vivo (1)H NMR study of pituitary adenomas to differentiate various immunohistochemical subtypes. Sci Rep. 2019;9:3007. https://doi.org/10.1038/s41598-019-38542-6.
Ijare OB, Holan C, Hebert J, Sharpe MA, Baskin DS, Pichumani K. Elevated levels of circulating betahydroxybutyrate in pituitary tumor patients may differentiate prolactinomas from other immunohistochemical subtypes. Sci Rep. 2020;10:1334. https://doi.org/10.1038/s41598-020-58244-8.
Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. https://doi.org/10.1186/s13059-017-1215-1.
Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. https://doi.org/10.1038/nrg2005.
Kober P, Boresowicz J, Rusetska N, Maksymowicz M. The role of aberrant DNA methylation in misregulation of gene expression in gonadotroph nonfunctioning pituitary tumors. Cancers (Basel). 2019;11(11):1650. https://doi.org/10.3390/cancers11111650.
Cheng S, Li C, Xie W, Miao Y, Guo J, Wang J, et al. Integrated analysis of DNA methylation and mRNA expression profiles to identify key genes involved in the regrowth of clinically non-functioning pituitary adenoma. Aging (Albany NY). 2020;12:2408–27. https://doi.org/10.18632/aging.102751.
Duong CV, Emes RD, Wessely F, Yacqub-Usman K, Clayton RN, Farrell WE. Quantitative, genome-wide analysis of the DNA methylome in sporadic pituitary adenomas. Endocr Relat Cancer. 2012;19:805–16. https://doi.org/10.1530/erc-12-0251.
Ling C, Pease M, Shi L, Punj V, Shiroishi MS, Commins D, et al. A pilot genome-scale profiling of DNA methylation in sporadic pituitary macroadenomas: association with tumor invasion and histopathological subtype. PLoS One. 2014;9:e96178. https://doi.org/10.1371/journal.pone.0096178.
Salomon MP, Wang X, Marzese DM, Hsu SC, Nelson N. The epigenomic landscape of pituitary adenomas reveals specific alterations and differentiates among acromegaly, Cushing’s disease and endocrine-inactive subtypes. Clin Cancer Res. 2018;24:4126–36. https://doi.org/10.1158/1078-0432.ccr-17-2206.
Crick F. Central dogma of molecular biology. Nature. 1970;227:561–3. https://doi.org/10.1038/227561a0.
Lee H, Zhang Z, Krause HM. Long noncoding RNAs and repetitive elements: Junk or intimate evolutionary partners? Trends Genet. 2019;35:892–902. https://doi.org/10.1016/j.tig.2019.09.006.
Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. https://doi.org/10.1038/nrc.2017.99.
Li X, Gianoulis TA, Yip KY, Gerstein M, Snyder M. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell. 2010;143:639–50. https://doi.org/10.1016/j.cell.2010.09.048.
Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun (Lond). 2018;38:27. https://doi.org/10.1186/s40880-018-0301-4.
Ahuja M, Jha A, Maléth J, Park S, Muallem S. cAMP and Ca2+ signaling in secretory epithelia: crosstalk and synergism. Cell Calcium. 2014;55:385–93. https://doi.org/10.1016/j.ceca.2014.01.006.
Spada A, Lania A, Mantovani S. Cellular abnormalities in pituitary tumors. Metabolism. 1996;45:46–8. https://doi.org/10.1016/s0026-0495(96)90079-7.
Lania A, Gil-del-Alamo P, Saccomanno K, Persani L, Faglia G, Spada A. Mechanism of action of pituitary adenylate cyclase-activating polypeptide (PACAP) in human nonfunctioning pituitary tumors. J Neuroendocrinol. 1995;7:695–702. https://doi.org/10.1111/j.1365-2826.1995.tb00811.x.
Romoli R, Lania A, Mantovani G, Corbetta S, Persani L, Spada A. Expression of calcium-sensing receptor and characterization of intracellular signaling in human pituitary adenomas. J Clin Endocrinol Metab. 1999;84:2848–53. https://doi.org/10.1210/jcem.84.8.5922.
Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–29. https://doi.org/10.1038/nrm1155.
Carafoli E, Santella L, Branca D, Brini M. Generation, control, and processing of cellular calcium signals. Crit Rev Biochem Mol Biol. 2001;36:107–260. https://doi.org/10.1080/20014091074183.
Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–19. https://doi.org/10.1038/nrg1879.
Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol. 2019;59:125–32. https://doi.org/10.1016/j.semcancer.2019.07.009.
Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: biological and therapeutic significance. Semin Cancer Biol. 2019;59:147–60. https://doi.org/10.1016/j.semcancer.2019.05.012.
Fattahi S, Amjadi-Moheb F, Tabaripour R, Ashrafi GH, Akhavan-Niaki H. PI3K/AKT/mTOR signaling in gastric cancer: epigenetics and beyond. Life Sci. 2020;262:118513. https://doi.org/10.1016/j.lfs.2020.118513.
Chamcheu JC, Roy T, Uddin MB, Banang-Mbeumi S, Chamcheu RN, Walker AL, et al. Role and therapeutic targeting of the PI3K/Akt/mTOR signaling pathway in skin cancer: a review of current status and future trends on natural and synthetic agents therapy. Cells. 2019;8:803. https://doi.org/10.3390/cells8080803.
Trovato M, Torre ML, Ragonese M, Simone A, Scarfì R, Barresi V, et al. HGF/c-met system targeting PI3K/AKT and STAT3/phosphorylated-STAT3 pathways in pituitary adenomas: an immunohistochemical characterization in view of targeted therapies. Endocrine. 2013;44:735–43. https://doi.org/10.1007/s12020-013-9950-x.
Murat CB, Braga PB, Fortes MA, Bronstein MD, Corrêa-Giannella ML, Giorgi RR. Mutation and genomic amplification of the PIK3CA proto-oncogene in pituitary adenomas. Braz J Med Biol Res. 2012;45:851–5. https://doi.org/10.1590/s0100-879x2012007500115.
Wang RQ, Lan YL, Lou JC, Lyu YZ, Hao YC, Su QF, et al. Expression and methylation status of LAMA2 are associated with the invasiveness of nonfunctioning PitNET. Ther Adv Endocrinol Metab. 2019;10:2042018818821296. https://doi.org/10.1177/2042018818821296.
Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–74. https://doi.org/10.1172/jci34739.
Xiao Z, Yang X, Zhang K, Liu Z, Shao Z, Song C, et al. Estrogen receptor α/prolactin receptor bilateral crosstalk promotes bromocriptine resistance in prolactinomas. Int J Med Sci. 2020;17:3174–89. https://doi.org/10.7150/ijms.51176.
Pópulo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. Int J Mol Sci. 2012;13:1886–918. https://doi.org/10.3390/ijms13021886.
Rubinfeld H, Shimon I. PI3K/Akt/mTOR and Raf/MEK/ERK signaling pathways perturbations in non-functioning pituitary adenomas. Endocrine. 2012;42:285–91. https://doi.org/10.1007/s12020-012-9682-3.
Jia W, Sanders AJ, Jia G, Liu X, Lu R, Jiang WG. Expression of the mTOR pathway regulators in human pituitary adenomas indicates the clinical course. Anticancer Res. 2013;33:3123–31.
Tanoue T, Nishida E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol Ther. 2002;93:193–202. https://doi.org/10.1016/s0163-7258(02)00188-2.
Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–7. https://doi.org/10.1016/s1470-2045(05)70168-6.
Yong HY, Koh MS, Moon A. The p38 MAPK inhibitors for the treatment of inflammatory diseases and cancer. Expert Opin Investig Drugs. 2009;18:1893–905. https://doi.org/10.1517/13543780903321490.
Peng WX, Huang JG, Yang L, Gong AH, Mo YY. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol Cancer. 2017;16:161. https://doi.org/10.1186/s12943-017-0727-3.
Ewing I, Pedder-Smith S, Franchi G, Ruscica M, Emery M, Vax V, et al. A mutation and expression analysis of the oncogene BRAF in pituitary adenomas. Clin Endocrinol (Oxf). 2007;66:348–52. https://doi.org/10.1111/j.1365-2265.2006.02735.x.
Roof AK, Gutierrez-Hartmann A. Consider the context: Ras/ERK and PI3K/AKT/mTOR signaling outcomes are pituitary cell type-specific. Mol Cell Endocrinol. 2018;463:87–96. https://doi.org/10.1016/j.mce.2017.04.019.
Pagotto U, Arzberger T, Theodoropoulou M, Grübler Y, Pantaloni C, Saeger W, et al. The expression of the antiproliferative gene ZAC is lost or highly reduced in nonfunctioning pituitary adenomas. Cancer Res. 2000;60:6794–9.
Theodoropoulou M, Zhang J, Laupheimer S, Paez-Pereda M, Erneux C, Florio T, et al. Octreotide, a somatostatin analogue, mediates its antiproliferative action in pituitary tumor cells by altering phosphatidylinositol 3-kinase signaling and inducing Zac1 expression. Cancer Res. 2006;66:1576–82. https://doi.org/10.1158/0008-5472.can-05-1189.
Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38:167–97. https://doi.org/10.1016/j.ccell.2020.06.001.
Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal Biochem. 2006;354:279–89. https://doi.org/10.1016/j.ab.2006.05.024.
Sabatino ME, Grondona E, Sosa LDV, Mongi Bragato B, Carreño L, Juarez V, et al. Oxidative stress and mitochondrial adaptive shift during pituitary tumoral growth. Free Radic Biol Med. 2018;120:41–55. https://doi.org/10.1016/j.freeradbiomed.2018.03.019.
Zhan X, Li J, Zhou T. Targeting Nrf2-mediated oxidative stress response signaling pathways as new therapeutic strategy for pituitary adenomas. Front Pharmacol. 2021;12:565748. https://doi.org/10.3389/fphar.2021.565748.
Zhan X, Desiderio DM. The use of variations in proteomes to predict, prevent, and personalize treatment for clinically nonfunctional pituitary adenomas. EPMA J. 2010;1:439–59. https://doi.org/10.1007/s13167-010-0028-z.
Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98. https://doi.org/10.1038/nrc3365.
Annesley SJ, Fisher PR. Mitochondria in health and disease. Cells. 2019;8(7):680. https://doi.org/10.3390/cells8070680.
Srinivasan S, Guha M, Kashina A, Avadhani NG. Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim Biophys Acta Bioenerg. 2017;1858:602–14. https://doi.org/10.1016/j.bbabio.2017.01.004.
Horoupian DS. Large mitochondria in a pituitary adenoma with hyperprolactinemia. Cancer. 1980;46:537–42. https://doi.org/10.1002/1097-0142(19800801)46:3%3c537::aid-cncr2820460319%3e3.0.co;2-6.
Saeger W, Lüdecke DK, Buchfelder M, Fahlbusch R, Quabbe HJ, Petersenn S. Pathohistological classification of pituitary tumors: 10 years of experience with the German Pituitary Tumor Registry. Eur J Endocrinol. 2007;156:203–16. https://doi.org/10.1530/eje.1.02326.
An J, Zhang Y, He J, Zang Z, Zhou Z, Pei X, et al. Lactate dehydrogenase A promotes the invasion and proliferation of pituitary adenoma. Sci Rep. 2017;7:4734. https://doi.org/10.1038/s41598-017-04366-5.
Dénes J, Swords F, Rattenberry E, Stals K, Owens M, Cranston T, et al. Heterogeneous genetic background of the association of pheochromocytoma/paraganglioma and pituitary adenoma: results from a large patient cohort. J Clin Endocrinol Metab. 2015;100:E531-541. https://doi.org/10.1210/jc.2014-3399.
Wang D, Wong HK, Feng YB, Zhang ZJ. 18beta-glycyrrhetinic acid induces apoptosis in pituitary adenoma cells via ROS/MAPKs-mediated pathway. J Neurooncol. 2014;116:221–30. https://doi.org/10.1007/s11060-013-1292-2.
Xiong S, Mu T, Wang G, Jiang X. Mitochondria-mediated apoptosis in mammals. Protein. Cell. 2014;5:737–49. https://doi.org/10.1007/s13238-014-0089-1.
Gao F, Pan S, Liu B, Zhang H. TFF3 knockout in human pituitary adenoma cell HP75 facilitates cell apoptosis via mitochondrial pathway. Int J Clin Exp Pathol. 2015;8:14568–73.
Tanase C, Albulescu R, Codrici E, Calenic B, Popescu ID, Mihai S, et al. Decreased expression of APAF-1 and increased expression of cathepsin B in invasive pituitary adenoma. Onco Targets Ther. 2015;8:81–90. https://doi.org/10.2147/ott.s70886.
Li N, Zhan X. Mitochondrial dysfunction pathway networks and mitochondrial dynamics in the pathogenesis of pituitary adenomas. Front Endocrinol (Lausanne). 2019;10:690. https://doi.org/10.3389/fendo.2019.00690.
Deyu H, Luqing C, Xianglian L, Pu G, Qirong L, Xu W, et al. Protective mechanisms involving enhanced mitochondrial functions and mitophagy against T-2 toxin-induced toxicities in GH3 cells. Toxicol Lett. 2018;295:41–53. https://doi.org/10.1016/j.toxlet.2018.05.041.
Kar S. Unraveling cell-cycle dynamics in cancer. Cell Syst. 2016;2:8–10. https://doi.org/10.1016/j.cels.2016.01.007.
Simpson DJ, Hibberts NA, McNicol AM, Clayton RN, Farrell WE. Loss of pRb expression in pituitary adenomas is associated with methylation of the RB1 CpG island. Cancer Res. 2000;60:1211–6.
Yoshino A, Katayama Y, Ogino A, Watanabe T, Yachi K, Ohta T, et al. Promoter hypermethylation profile of cell cycle regulator genes in pituitary adenomas. J Neurooncol. 2007;83:153–62. https://doi.org/10.1007/s11060-006-9316-9.
García-Fernández RA, García-Palencia P, Sánchez M, Gil-Gómez G, Sánchez B, Rollán E, et al. Combined loss of p21(waf1/cip1) and p27(kip1) enhances tumorigenesis in mice. Lab Invest. 2011;91:1634–42. https://doi.org/10.1038/labinvest.2011.133.
Gruppetta M, Formosa R, Falzon S, Ariff Scicluna S, Falzon E, Degeatano J, et al. Expression of cell cycle regulators and biomarkers of proliferation and regrowth in human pituitary adenomas. Pituitary. 2017;20:358–71. https://doi.org/10.1007/s11102-017-0803-0.
Matoušek P, Buzrla P. Factors That predict the growth of residual nonfunctional pituitary adenomas: correlations between relapse and cell cycle Markers. Biomed Res Int. 2018;2018:1876290. https://doi.org/10.1155/2018/1876290.
Butz H, Németh K, Czenke D, Likó I, Czirják S, Zivkovic V, et al. Systematic investigation of expression of G2/M transition genes reveals CDC25 alteration in nonfunctioning pituitary adenomas. Pathol Oncol Res. 2017;23:633–41. https://doi.org/10.1007/s12253-016-0163-5.
Vázquez-Borrego MC, Fuentes-Fayos AC, Herrera-Martínez AD, F LL, Ibáñez-Costa A, Moreno-Moreno P, et al. Biguanides exert antitumoral actions in pituitary tumor cells through AMPK-dependent and -independent mechanisms. J Clin Endocrinol Metab. 2019; 104:3501–3513. https://doi.org/10.1210/jc.2019-00056.
Vázquez-Borrego MC, Fuentes-Fayos AC, Herrera-Martínez AD, Venegas-Moreno E, F LL, Fanciulli A, et al. Statins directly regulate pituitary cell function and exert antitumor effects in pituitary tumors. Neuroendocrinology. 2020; 110:1028–1041. https://doi.org/10.1159/000505923.
Zeng J, See AP, Aziz K, Thiyagarajan S, Salih T, Gajula RP, et al. Nelfinavir induces radiation sensitization in pituitary adenoma cells. Cancer Biol Ther. 2011;12:657–63. https://doi.org/10.4161/cbt.12.7.17172.
Hubina E, Nanzer AM, Hanson MR, Ciccarelli E, Losa M, Gaia D, et al. Somatostatin analogues stimulate p27 expression and inhibit the MAP kinase pathway in pituitary tumours. Eur J Endocrinol. 2006;155:371–9. https://doi.org/10.1530/eje.1.02213.
Guzzo MF, Carvalho LR, Bronstein MD. Ketoconazole treatment decreases the viability of immortalized pituitary cell lines associated with an increased expression of apoptosis-related genes and cell cycle inhibitors. J Neuroendocrinol. 2015;27:616–23. https://doi.org/10.1111/jne.12277.
Petiti JP, Sosa LDV, Picech F, Moyano Crespo GD, Arevalo Rojas JZ, Pérez PA, et al. Trastuzumab inhibits pituitary tumor cell growth modulating the TGFB/SMAD2/3 pathway. Endocr Relat Cancer. 2018;25:837–52. https://doi.org/10.1530/erc-18-0067.
Lu M, Wang Y, Zhan X. The MAPK pathway-based drug therapeutic targets in pituitary adenomas. Front Endocrinol (Lausanne). 2019;10:330. https://doi.org/10.3389/fendo.2019.00330.
Dworakowska D, Wlodek E, Leontiou CA, Igreja S, Cakir M, Teng M, et al. Activation of RAF/MEK/ERK and PI3K/AKT/mTOR pathways in pituitary adenomas and their effects on downstream effectors. Endocr Relat Cancer. 2009;16:1329–38. https://doi.org/10.1677/erc-09-0101.
Xu M, Shorts-Cary L, Knox AJ, Kleinsmidt-DeMasters B, Lillehei K, Wierman ME. Epidermal growth factor receptor pathway substrate 8 is overexpressed in human pituitary tumors: role in proliferation and survival. Endocrinology. 2009;150:2064–71. https://doi.org/10.1210/en.2008-1265.
Rubinfeld H, Kammer A, Cohen O, Gorshtein A, Cohen ZR, Hadani M, et al. IGF1 induces cell proliferation in human pituitary tumors - functional blockade of IGF1 receptor as a novel therapeutic approach in non-functioning tumors. Mol Cell Endocrinol. 2014;390:93–101. https://doi.org/10.1016/j.mce.2014.04.007.
Sirotnak FM, Zakowski MF, Miller VA, Scher HI, Kris MG. Efficacy of cytotoxic agents against human tumor xenografts is markedly enhanced by coadministration of ZD1839 (Iressa), an inhibitor of EGFR tyrosine kinase. Clin Cancer Res. 2000;6:4885–92.
Fukuoka H, Cooper O, Mizutani J, Tong Y, Ren SG, Bannykh S, et al. HER2/ErbB2 receptor signaling in rat and human prolactinoma cells: strategy for targeted prolactinoma therapy. Mol Endocrinol. 2011;25:92–103. https://doi.org/10.1210/me.2010-0353.
Vlotides G, Siegel E, Donangelo I, Gutman S, Ren SG, Melmed S. Rat prolactinoma cell growth regulation by epidermal growth factor receptor ligands. Cancer Res. 2008;68:6377–86. https://doi.org/10.1158/0008-5472.can-08-0508.
Zatelli MC, Minoia M, Filieri C, Tagliati F, Buratto M, Ambrosio MR, et al. Effect of everolimus on cell viability in nonfunctioning pituitary adenomas. J Clin Endocrinol Metab. 2010;95:968–76. https://doi.org/10.1210/jc.2009-1641.
Cerovac V, Monteserin-Garcia J, Rubinfeld H, Buchfelder M, Losa M, Florio T, et al. The somatostatin analogue octreotide confers sensitivity to rapamycin treatment on pituitary tumor cells. Cancer Res. 2010;70:666–74. https://doi.org/10.1158/0008-5472.can-09-2951.
Koklesova L, Samec M, Liskova A, Zhai K, Büsselberg D, Giordano FA, Kubatka P, Golunitschaja O. Mitochondrial impairments in aetiopathology of multifactorial diseases: common origin but individual outcomes in context of 3P medicine. EPMA J. 2021;12(1):1–14. https://doi.org/10.1007/s13167-021-00237-2.
Acknowledgements
The authors acknowledge the financial support from the Shandong First Medical University Talent Introduction Funds (to X.Z.), Shandong First Medical University High-level Scientific Research Achievement Cultivation Funding Program (to X.Z.), the Shandong Provincial Natural Science Foundation (ZR202103020356/ZR2021MH156 to X.Z.), and the Academic Promotion Program of Shandong First Medical University (2019ZL002).
Funding
This work was supported by the Shandong First Medical University Talent Introduction Funds (to X.Z.), Shandong First Medical University High-level Scientific Research Achievement Cultivation Funding Program (to X.Z.), the Shandong Provincial Natural Science Foundation (ZR202103020356/ ZR2021MH156 to X.Z.), and the Academic Promotion Program of Shandong First Medical University (2019ZL002).
Author information
Authors and Affiliations
Contributions
S.Q. collected and analyzed literature, and wrote the manuscript. C.L. participated in the collection and analysis of literature. X.Z. conceived the concept, designed the manuscript, coordinated, wrote, and critically revised the manuscript, and was responsible for its financial support and the corresponding works. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wen, S., Li, C. & Zhan, X. Muti-omics integration analysis revealed molecular network alterations in human nonfunctional pituitary neuroendocrine tumors in the framework of 3P medicine. EPMA Journal 13, 9–37 (2022). https://doi.org/10.1007/s13167-022-00274-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-022-00274-5
Keywords
- Nonfuctional pituitary neuroendocrine tumor (NF-PitNET)
- Invasive NF-PitNET
- Aggressive NF-PitNET
- Multi-omics integration analysis
- Calcium signaling pathway
- cGMP-PKG signaling pathway
- mTOR signaling pathway
- PI3K/AKT signaling pathway
- MAPK signaling pathway
- Oxidative stress response
- Mitochondrial dysfunction
- Cell cycle dysregulation
- Signaling pathway
- Molecular network
- Biomarker
- Therapeutic target
- Patient stratification
- Predictive diagnosis
- Prognostic assessment
- Targeted therapy
- Predictive preventive personalized medicine (3P medicine; PPPM)