Abstract
Background
Colorectal cancer has high incidence and mortality rates. The signal transducer and activator of transcription (STAT) family plays vital roles in the tumorigenesis and development of colorectal cancer. The expression, prognostic value, and immune function of the STAT family are becoming much more clearly.
Methods
Our study collected data from several public data portals such as TCGA (644 samples) and GTEx database (308 samples) and clinical samples (30 samples, China). Then we systematically assessed the expression level and prognostic value of the STAT family in colorectal cancer samples. Moreover, the immune function and immune infiltration levels of prognosis-related STAT members were explored via single cell RNA-seq and spatial transcriptomics technology data. Several useful portals and tools have been utilized such as CancerSEA and TISIDB in single-cell analysis, CBio Cancer Genomics in multidimensional alterations, MethSurv in DNA methylation, and related R packages.
Results
Our study found that STAT3 and STAT5B were significantly lower in colorectal cancer via multi-omics (P < 0.001). Higher STAT3 and STAT5B level were correlated with better future outcome. Nomograms were developed to predict the distal survival time (C-index = 0.724). The functions of STAT3 and STAT5B are associated with inflammation, the JAK/STAT pathway and the immune response. The major cell types of colorectal cancer were CD4Tconv, CD8T, CD8Tex, Tprolif, Treg and STAT3 and STAT5B widely expressed in these cells. STAT3 and STAT5B both correlated with CD244 and KDR for immune checkpoints.
Conclusion
STAT3 and STAT5B are downregulated in colorectal cancer and have great potential as prognostic biomarkers and novel immunotherapy targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Worldwide, colorectal cancer was the second leading cause of cancer-related mortality and the third most prevalent cancer now, representing a huge global health burden [1]. Although treatment schedules such as surgery, systemic chemotherapy and even stratified medicine approaches have become increasingly optimized, the survival rate of colorectal cancer is still low [2]. Immune check point related treating has greatly changed the therapeutic strategy but it is not as robust as in colorectal cancer compared as other tumours [3,4,5]. Thus, it is necessary to explore useful biomarkers for prognosis prediction and monitoring as well as novel treatment targets.
The signal transducer and activator of transcription (STAT) protein family contains seven members, including STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6, which play vital roles in the cell cycle, immune responses and tumour development by altering gene transcription in effector cells [6]. A growing body of studies have suggested that STATs are extensively dysregulated in several cancers, especially colon cancer [7], gastric cancer [8] and hepatic carcinoma [9], thus showing their potential for tumour screening, dynamic monitoring and further therapy.
More and more evidence pointed out that STAT mediates large tumor-driven immune regulatory processes and sustained inflammation such as STAT3 in colorectal cancer [10]. The current study aims to analyse the expression features of the STAT family and identify two prognostic biomarkers to help further optimize immunotherapy.
2 Methods
2.1 Public database retrieval and data processing
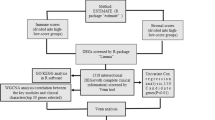
Clinical and pathological information of colorectal cancer and the RNA-seq data of 644 CRC tumours and para-carcinoma tissues generated by The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/tcga) and 308 normalized tissues’ RNA-seq data (version 7) from the Genotype-Tissue Expression (GTEx) data portal (https://www.gtexportal.org/home/index.html) were downloaded [11]. Clinical colorectal cancer tissue samples used in our study were obtained from the First Affiliated Hospital of Ningbo University, China, between 2022 and 2023. Colorectal cancer tissues and paired the adjacent normal tissues (5 cm away from the edge of the tumour) were collected from 30 patients underwent surgical procedures. Two human colorectal cancer cell lines (HCT116, HT29) and the colon cell line NCM460 were obtained from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, China. All of these patients have signed informed consents and this study was approved by the Ethics Committee of the Affiliated Hospital of Medical School of Ningbo University (No. KY20210762).
2.2 Distinguishingly expressed and prognostic STAT identification
This study first identified the differentially expressed STATs in colorectal cancer from the TCGA cohort and normal tissues in the GTEx cohort using the R statistical computing environment (3.6.3; R Foundation for Statistical Computing). A univariate Cox regression model was used to screen the differentially expressed STATs that are correlated with the overall survival (OS) time and disease-specific survival (DSS) time of the TCGA colorectal training cohort. STATs with a p value less than 0.05 were included in the multivariate Cox regression model [12]. STATs were considered to be prognostic genes if the p values were less than 0.1 in the multivariate Cox regression model. MethSurv (https://biit.cs.ut.ee/methsurv/) is a web tool to perform multivariable survival analysis using DNA methylation data which was obtained from TCGA methylation data profiled using HM450K array. The relationship between DNA methylation level and expression level was detected using MethSurv tool [13]
2.3 Survival analysis and nomogram of prognosis-related STATs
Then, the survival data from TCGA database was analysed using the Kaplan–Meier method via the R package survival v 3.2–10 [14]. Some common clinical characteristics, including age, BMI, CEA level, history of colon polyps, colon polyps present and the expression level of prognosis-related genes, were selected to construct a nomogram to predict the 3- and 5 year OS of colorectal patients. Additionally, the concordance index (C‐index) was used to assess the discrimination of the nomogram, and calibration plots were used to show the association between the predicted and observed risk results [15].
2.4 Single-cell analysis of prognosis-related STATs
CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp), the first database devoted to comprehensively decoding the functional states of cancer cells at a single-cell resolution, was applied to explore the function of STAT3 and STAT5B in the cell line [16]. Sparkle v1.0 (https://grswsci.top/analyze/) and SpatialTME (https://www.spatialtme.yelab.site/) are online tools of spatial transcriptomics research where we can use scRNAseq data to deconvolute the cell types captured and visualize the spatial relationship of the individual datapoints of scRNA-seq via Cottrazm package [17, 18].
2.5 Functional analysis of prognosis-related STATs
The CBio Cancer Genomics Portal (http://cbioportal.org) was used to explore multidimensional alterations of STAT3 and STAT5B in TCGA colorectal samples [19]. Gene set enrichment analysis was conducted using the MSigDB Collections via R (version 3.6.3) for data processing and visualization, and |ES|> 1, P < 0.05, and FDR < 0.25 were considered to be statistically significant. Signatures with < 10 genes or > 500 genes were filtered out. [20, 21]. Metascape (http://metascape.org), a portal site for gene annotation and enrichment analysis, was used to perform KEGG pathway enrichment analysis and gene ontology (GO) classification. A p value < 0.05, min enrichment > 3, and min overlap > 3 were considered significant [22]. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) website (https://string-db.org/), containing various protein correlation data, was used to establish the STAT3-STAT5B interacting protein–protein interaction (PPI) network. A confidence score > 0.7 was considered significant [23].
2.6 Immunoinhibitory analysis
TISIDB (http://cis.hku.hk/TISIDB/index.php), a web portal integrating multiple heterogeneous data types, was used for detecting tumour and immune system interactions [24]. The relations between STAT3, STAT5B and immunoinhibitors were explored in this database. A p value < 0.05 was considered significant.
2.7 Immune infiltration analysis
We explored the associations between these prognosis-related genes and the abundance of infiltrating immune cells in colon cancer and rectal cancer, such as CD4+ T cells, CD8+ T cells, cancer-associated fibroblasts (CAFs), myeloid-derived suppressor cells (MDSCs), and natural killer cells (NK cells), using the Tumor IMmune Estimation Resource 2.0 (http://timer.cistrome.org/) [25,26,27]. Furthermore, the estimated score, stromal score and immune score were computed to estimate the infiltration degree of immune cells and stromal cells via the R package ESTIMATE v1.0.13 using the default parameters [28]. Pearson's correlation analysis was conducted to determine the correlation between the expression of selective genes and the three scores. Tumor Immunization Single Cell Center (TISCH, http://tisch.comp-genomics.org/home/) is a single-cell RNA sequencing (scRNA-seq) database about various tumour microenvironment, which was performed to detect the purity and immune infiltration of colorectal cancer [29].
2.8 Expression validation of prognosis-related STATs
All cell and tissue RNA were extracted by TRIzol (Ambion, Carlsbad, USA) and reversed transcribed to cDNA with a GoScript Reverse Transcription (RT) System (Promega, Madison, USA). Quantitative real-time polymerase chain reaction (qRT-PCR) detection was performed with GoTaq qPCR Master Mix (Promega) whose conditions were as follows: 95 °C for 5 min, followed by 40 cycles of 94 °C for 15 s, 50 °C for 30 s, and 72 °C for 30 s. GAPDH mRNA was chosen to normalize and the primer sequences were as follows: STAT3: forward, 5′- CCCCGTACCTGA AGACCAAG-3′, reverse, 5′-TCCTCACATGGGGGAGGTAG-3′; STAT5B: forward, 5′-GGATGGGTGCATCGG GGAAG-3′, reverse, 5′- TCT CAG AGG CAG GTG CTG GT-3′;GAPDH: forward, 5′-ACCCACTCCTCCACCTTTGAC-3′, reverse, 5′-TGTTGCTGTAGCCAAATTCGTT-3′. ΔCt method was used to quantify (ΔCt = Ctgene–CtGAPDH). A higher ΔCt value means a lower level [30]. Immunohistochemistry results were acquired from the Human Protein Atlas (https://www.proteinatlas.org/).
3 Results
3.1 Inconsistent expression levels of STATs in colorectal cancer
As shown in Fig. 1, the mRNA levels of STAT2 (n = 4.85Tumor vs 5.80normal), STAT3 (n = 5.63Tumor vs 6.03normal), STAT4 (n = 0.95Tumor vs 1.47normal), STAT5A (n = 3.83Tumor vs 4.95normal), STAT5B (n = 4.12Tumor vs 5.03normal), and STAT6 (n = 6.41Tumor vs 6.99normal) were all downregulated in colorectal cancer tissues compared to normal samples but upregulated in STAT1 (n = 5.86Tumor vs 4.69normal). We gathered and consolidated these data and displayed the expression distribution of the STAT family between colorectal cancer and normal controls in the TCGA and GTEx cohorts using the heatmap in Fig. 1H.
The mRNA expression levels of STAT family members from the TCGA and GTEx databases. A The mRNA expression of STAT1 was notably higher in colorectal cancer. B The mRNA expression of STAT2 was notably lower in colorectal cancer. C The mRNA expression of STAT3 was notably lower in colorectal cancer. D The mRNA expression of STAT4 was notably lower in colorectal cancer. E The mRNA expression of STAT5A was notably lower in colorectal cancer. F The mRNA expression of STAT5B was notably lower in colorectal cancer. G The mRNA expression of STAT6 was notably lower in colorectal cancer. H The distribution of STAT family expression between colorectal cancer and normal controls in the TCGA and GTEx cohorts. Downregulated represented by a blue color and upregulated represented by a red colour (*P < 0.05; ** P < 0.01; *** P < 0.001)
The clinical information about 322 colorectal patients of the TCGA cohort is divided according to the median expression of STATs shown in Tables 1, 2 using chi-square analysis. Obviously, lower expression of STAT3 was related to much older age (P = 0.002), lower risk of perineural invasion (P = 0.022) and bad progression-free interval (P = 0.011). Likewise, lower risk of STAT5B correlated to much older age (P = 0.003), higher T stage (P < 0.001) and colon polyps history (P = 0.005).
There was a significant correlation between the DNA methylation level and mRNA expression level among STAT3 and STAT5B, which implied that the inconsistent expression level of STATs may be associated with aberrant DNA methylation, as shown in Supplementary Fig. 1 (Fig. S1).
3.2 Two STATs with prognostic value for colorectal cancer
Based on univariate and multivariate Cox regression analyses, STAT3 (HR 0.658; P = 0.077) and STAT5B (HR 0.667; P = 0.083) were significant predictors of OS (Table 3). Additionally, STAT5B significantly predicted DSS (HR 0.647; P = 0.015, Table 4). Both were independent prognostic biomarkers for colorectal cancer. In addition, the Kaplan–Meier curve showed that higher STAT3 levels were correlated with increased OS (P = 0.031) and DSS (P = 0.016) in colorectal cancer patients (Fig. 2A, B). Elevated STAT5B levels were also associated with increased OS (P = 0.015) and DSS (P = 0.011) in colorectal cancer patients (Fig. 2C, D).
Survival difference between the expression of STAT3 and STAT5B and colorectal cancer from TCGA. A Low STAT3 expression was associated with poor overall survival time in colorectal cancer patients. B Patients with lower STAT3 expression were associated with poor disease-specific survival time in colorectal cancer. C Low STAT5B expression was associated with poor overall survival time in colorectal cancer patients. D Low STAT5B expression was associated with poor disease-specific survival time in colorectal cancer
3.3 Establishment of nomogram
Given the favourable prognostic value of STAT3 and STAT5B, we combined these genes with several significant clinicopathologic characteristics from Tables 1, 2 and built a prognostic prediction nomogram to predict the 3- and 5-year OS (Fig. 3A). The C-index for OS was 0.724 (95% CI 0.675–0.772). The calibration curves displayed a satisfying consensus between the OS prediction by the nomogram and actual observation at 3 and 5 years, as shown in Figs. 3B, C, which will be a practical tool to predict distal survival time for CRC patients.
3.4 Functional analysis of STAT3 and STAT5B
CancerSEA was firstly performed to demonstrate the function of STAT3 and STAT5B in the cell line. Our results suggested that STAT3 levels correlated with hypoxia and stemness in colorectal cancer cells (Fig. 4A). Then, GSEA was used to investigate the biological function and signalling pathways of STAT3 and STAT5B in colorectal cancer samples. According to the MSigDB Collections, GSEA revealed several signal pathways from STAT3 and STAT5B expression datasets, as shown in Supplementary Table 1 and Fig. 4B, C. G protein-coupled receptor ligand binding and neutrophil degranulation were differentially enriched in the STAT3 expression phenotype, while the olfactory signalling pathway and metabolism of amino acids and derivatives were remarkably enriched in the STAT5B expression phenotype. KEGG pathway enrichment analysis and gene ontology (GO) classification further annotated the functions of STAT3 and STAT5B in colorectal cancer samples, as shown in Supplementary Table 2–3 and Fig. 4D, E. Interestingly, the function of STAT3 was focused on chromatin assembly and DNA packaging, which implied that STAT3 may disturb the progression of gene expression and chromatin assembly in colorectal cancer. STAT5B may influence the humoral response, signal transduction and receptor ligand activity during the development of colorectal cancer. The gene network also proves that STAT3 and STAT5B are strongly correlated with inflammation, the JAK/STAT pathway and the immune response (Fig. 5A). The PPI network displays the proteins interacting with STAT3 and STAT5B, such as JAKs, IL10RA, and EGFR (Fig. 5B).
Biological function analysis of STAT3 and STAT5B. A Single-cell function analysis from CancerSEA showed that STAT3 was associated with hypoxia and stemness in colorectal cancer cells. B GSEA of STAT3 in the colorectal cancer cohort from TCGA. C GSEA of STAT5B in the colorectal cancer cohort from TCGA. D KEGG and GO enrichment analyses of coexpressed genes of STAT3. E KEGG and GO enrichment analyses of coexpressed genes of STAT5B
Gene and protein interaction diagrams. A The gene network associated with STAT3 and STAT5B using GeneMANIA. The functions of the nodes were represented by different colour part in the circle. The correlations between nodes were represented by different colour of networks. B PPI network diagram of interactions between proteins encoded by the STAT3 and STAT5B genes from STRING. Edges represent protein–protein associations via different colours. Purple, blue, yellow, green, pink respectively represent gene co-expression, gene co-occurrence, gene textmining, gene neighbourhood, gene experimentally determined
3.5 Associations between STAT3, STAT5B and immune inhibitors
Recently, immune checkpoint inhibitors have shown remarkable progress in the treatment of advanced cancer and improving distant prognosis [31, 32]. Thus, we evaluated the association of the expression of STAT3 and STAT5B with immune checkpoint inhibitors in colon cancer and rectal cancer via the TISIDB database. Our results indicated that STAT3 was simultaneously associated with ADORA2A, BTLA, CD96, CD244, CD274, CSF1R, CTLA4, IDO1, KDR, LAG3, LGALS9, PDCD1, PDCD1LG2, and TIGIT in colon and rectal cancer. STAT5B was correlated with CD244 and KDR in colon and rectal cancer (Fig S2).
3.6 Correlations of STAT3 and STAT5B with immune infiltration
In light of the vital role of immune cells in the microenvironment and the association between STATs and the immune response confirmed by GSEA, KEGG and GO analysis as reported before, we further explored the correlation between STAT expression and immune infiltration. Our results showed that the expression level of STAT3 was positively correlated with CD4+ T cells, CD8+ T cells, and cancer-associated fibroblasts but negatively correlated with purity and MDSCs in colorectal cancer based on the TIMER 2.0 database (Fig. 6A, B). Furthermore, the expression level of STAT5B was negatively associated with tumour purity and MDSCs but positively associated with CD4+ T cells, CD8+ T cells, cancer-associated fibroblasts and NK cells in colon cancer (Fig. 6C). The expression level of STAT5B was negatively associated with tumour purity, NK cells and MDSCs but positively associated with CD4+ T cells, CD8+ T cells and cancer-associated fibroblasts in rectal cancer (Fig. 6D). The correlations of STAT3, STAT5B and immune cell markers are shown in Supplementary Tables 4–5.
Immune cell infiltration levels and scores. A STAT3 was positively correlated with CD4 + T cells, CD8 + T cells, and cancer-associated fibroblasts but negatively correlated with purity and MDSCs in colon cancer. B STAT3 was positively correlated with CD4 + T cells, CD8 + T cells, and cancer-associated fibroblasts but negatively correlated with purity and MDSCs in rectal cancer. C STAT5B was negatively associated with tumour purity and MDSCs but positively associated with CD4 + T cells, CD8 + T cells, cancer-associated fibroblasts and NK cells in colon cancer. D STAT5B was negatively associated with tumour purity, NK cells and MDSCs but positively associated with CD4 + T cells, CD8 + T cells and cancer-associated fibroblasts in rectal cancer. E Higher expression of STAT3 was associated with higher ESTIMATE scores, immune scores and stromal scores. F Higher STAT5B expression was associated with a higher ESTIMATE Score and Stromal Score
Moreover, the STROMAL score, IMMUNE score, and ESTIMATE score for each colorectal cancer sample were computed from TCGA via the ESTIMATE method. Spearman correlation analysis showed that the expression of STAT3 was positively related to the IMMUNE score, STROMAL score and ESTIMATE score (Fig. 6E). The expression of STAT5B was positively correlated with STROMAL score and ESTIMATE score (Fig. 6F). The expression of STAT3 and STAT5B in the microenvironment of colorectal cancer was analyzed as well. The major cell types of colorectal cancer were CD4Tconv, CD8T, CD8Tex, Tprolif, Treg (Fig S3A-B). The STAT3 and STAT5B widely expression in these cells (Fig S3C-D). Spatial transcriptomics quantificated and visualized of STAT3 annd STAT5B expression patterns in individual tissue sections and regions in Fig. 7A, D. More interestingly, the correlations of expression level of STAT3 and STAT5B and tumour microenvironment were annotated in Fig. 7E, F. All these data suggest the crucial role of STAT3 and STAT5B in the immune regulation of colorectal cancer.
The landscape of STAT3 and STAT5B in tumor microenvironment via spatial transcriptomics. A The expression level of STAT3 in different positions of colorectal cell. B Localization of immune cells with the highest content in each spot according to the spatial transcriptome deconvolution of STAT3. C The expression level of STAT5B in different positions of colorectal cell. D Localization of immune cells with the highest content in each spot according to the spatial transcriptome deconvolution of STAT5B. E The correlation between immune cell counts and expression level of STAT3 in all spots via spearman correlation analysis calculation. F The correlation between immune cell counts and expression level of STAT5B in all spots via spearman correlation analysis calculation
3.7 Validation of the differential expression of STAT3 and STAT5B
Finally, we compared the expression level of STAT3 and STAT5B in the cell line and tissue line by qRT-PCR for validation. The results revealed that STAT3 and STAT5B expression were prominently lower in the colorectal cancer cells (Fig. 8A, B) and tissues (Fig. 8C, D), which was consistent with the previous studies. Moreover, the immunohistochemistry results were shown in Fig S4. All of this evidence supported our results.
The expression of STAT3 and STAT5B via qRT-PCR. A STAT3 was downregulated in colorectal cells compared to the normal cell. B STAT5B was downregulated in colorectal cells compared to the normal cell. C STAT3 was significantly downregulated in colorectal tissues compared to the paired the adjacent normal tissues. D STAT5B was significantly downregulated in colorectal tissues compared to the paired the adjacent normal tissues
4 Discussion
STAT family members play vital roles in regulating gene expression and responding to signals from a variety of stimuli, and they have been proven to be master regulators of carcinogenesis in various cancers, thus indicating their potential as novel biomarkers and therapeutic targets of tumours [33]. It has been reported that the STAT family is a promising therapeutic and prognostic target in glioblastoma, but the function and expression of the STAT family in colorectal cancer remained unknown prior to the current study.
Our study first systemically explores the expression and prognostic and immunotherapy values of STAT family members in colorectal cancer. Obviously, the expression level of STATs is imbalanced in colorectal cancer, and both STAT3 and STAT5 are remarkably downregulated in cancer, which may be caused by abnormal DNA methylation. Univariate Cox analysis showed that STAT3 and STAT5B can have prognostic value for colorectal cancer from the TCGA cohort. Hence, the Kaplan–Meier curve was constructed, and our results showed that lower STAT3 and STAT5B levels are correlated with decreased OS and DSS in colorectal cancer, which is consistent with previous results. These findings indicate the potential of the STAT family as novel prognostic biomarkers.
Subsequently, a nomogram of OS was designed for prognostic models and optimize the efficacy of these biomarkers. We incorporated some common clinicopathologic characteristics to construct the model, and calibration curves showed a satisfying consensus. Certainly, the limitations of the current models were limited to a relatively small sample size. These models possess more precise effects for predicting the outcomes of patients with colorectal cancer, and they should be examined in larger clinical cohorts for further validation.
An increasing number of studies have focused on the unique role of the JAK/STAT pathway in cancer. STATs are mainly involved in tumour cell recognition, tumour microenvironment regulation and immune escape [34]. The JAK/STAT axis contributes to the progression and metastatic development of cervical cancer [35]. It has been investigated that CRH can promote human colon cancer cell proliferation via decreased STAT3 by IL-6/JAK2/STAT3 signal pathway and VEGF-induced tumor angiogenesis [36]. Huang’s research indicates that IGF2BP3 can activate the JAK/STAT pathway, further critically facilitating cell proliferation and cell cycle progression while inhibiting cell apoptosis in bladder cancer [37]. Hence, the function of single cells was assessed and GSEA was performed to annotate the function of prognostic genes. Our study displays the function of STAT3 and STAT5B in several dimensions and finds that STAT3 is correlated with hypoxia and stemness in cell lines. Next, GSEA, KEGG and GO analysis were applied for enrichment analysis, which implies that STAT3 is strongly associated with the activity of G protein-coupled receptors, neutrophil degranulation and chromatin assembly, while STAT5B is correlated with biological metabolic processes and humoral responses. The mutual function between associated proteins is especially necessary for annotating the molecular mechanism of carcinogenesis. The gene network and PPI network further enriched the synergistic genes and proteins of STAT3 and STAT5B. All these results show that STAT3 and STAT5B play critical roles in gene expression, immune response, inflammation, and JAK/STAT pathway activities in colorectal cancer. Moreover, recent study reveals ETHE1 can promote the interaction between T cell protein tyrosine phosphatase (TC45) and STAT3, resulting in decreased STAT3 phosphorylation and inhibition of the STAT3 signal pathway, which exhibits poorer prognosis of CRC patients [38]. STAT5B was found to be involved in cell growth, cell cycle progression and apoptosis of CRC cells via reduction of mitochondrial membrane potential and generation of reactive oxygen species, playing an important role of CRC tumorigenesis [39]. Th potential mechanisms behind the downregulation of STAT3 and STAT5B in CRC are gradually clarified.
Recently, checkpoint blockade immunotherapy has become much more promising for these advanced and drug-resistant cancers. The expression level of immune check-point molecules is a major mechanism for immune escape and immune response, which is vital for designing immune checkpoint blockade drugs [40, 41]. Based on previous results, the function of STATs is associated with the immune response. Therefore, we used TISIDB and TIMER to explore the immune landscape of STAT3 and STAT5B. We found that CD244 and KDR are the consensus sites between STAT3, STAT5B and colorectal cancer, which provides two potential targets for developing immune checkpoint drugs.
Moreover, tumour factors shape the tumour microenvironment and mediate the level and functions of tumour-infiltrating immune cells, which further influences the therapeutic effect and distal survival time [42, 43]. We investigated the infiltration levels of several kinds of immune cells. Our results indicate that lower STAT3 and STAT5B levels are as-sociated with lower CD4+ T cells, CD8+ T cells, and CAFs as well as higher MDSC numbers in colorectal cancer. CD4+ T cells and CD8+ T cells are the major tumour-antagonizing cells [44]. Lower STAT3 and STAT5B levels are associated with the downregulation of these cells, further promoting the environment of immune inhibition. Likewise, CAFs exhibit senescence-like phenotypes in the tumour microenvironment, and our results suggest that lower STAT3 and STAT5B levels may promote cell proliferation and inhibit apoptosis [45]. Furthermore, MDSCs can release immune suppressive factors to inhibit the immunological competence of T cells, indicating that lower STAT3 and STAT5B upregulate the level of MDSCs and ultimately decrease the amount of CD4+ T cells and CD8+ T cells [46]. The ESTIMATE score methods also reveal the correlation of these genes in immune regulation. These effects are consistent with the immune functions of STAT3 and STAT5B we identified before, which is worthy of further investigation.
To sum up, our study innovatively used multi-omics analyses to detect STAT family expression level and filtered key genes in colorectal cancer. Moreover, we systematically accessed their biological functions and clinical application values. Despite a robust data set, this study has several limitations. For example, the data of several colorectal cancer in some databases were not counted separately or the number was insufficient. Nevertheless, we cannot obtain adequate clinical samples covering all ages as the expression of STAT3 and STAT5B associated with ages. As two key prognostic-related genes have been filtered, further exploration of the correlations between two genes and underlying molecular or cellular mechanisms driving the observed effects should be pursued in the future. Meanwhile, a larger cohort would allow for more robust subgroup analyses and the identification of potential biomarkers or patient-specific responses.
5 Conclusion
In conclusion, STAT3 and STAT5B are downregulated in colorectal cancer with critical functions in immune response and inflammation, which have great potential as prognostic biomarkers and novel immunotherapy targets.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- CAFs:
-
Cancer-associated fibroblasts
- C-index:
-
Concordance index
- DSS:
-
Disease-specific survival
- GO:
-
Gene ontology
- GTEx:
-
Genotype-tissue expression
- MDSCs:
-
Myeloid-derived suppressor cells
- NK:
-
Natural killer
- OS:
-
Overall survival
- PPI:
-
Protein–protein interaction
- STAT:
-
Signal transducer and activator of transcription
- STRING:
-
Search tool for the retrieval of interacting genes/proteins
- TCGA:
-
The cancer genome atlas
References
Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63.
Patel SG, Dominitz JA. Screening for colorectal cancer. Ann Intern Med. 2024. https://doi.org/10.7326/AITC202404160.
Jiang C, Zhou Q, Yi K, et al. Colorectal cancer initiation: understanding early-stage disease for intervention. Cancer Lett. 2024;589: 216831.
Yan S, Wang W, Feng Z, et al. Immune checkpoint inhibitors in colorectal cancer: limitation and challenges. Front Immunol. 2024;15:1403533.
Gou H, Liu S, Liu L, et al. Obeticholic acid and 5β-cholanic acid 3 exhibit anti-tumor effects on liver cancer through CXCL16/CXCR6 pathway. Front Immunol. 2022;13:1095915.
Jamroskovic J, Doimo M, Chand K, et al. Quinazoline ligands induce cancer cell death through selective STAT3 inhibition and G-quadruplex stabilization. J Am Chem Soc. 2020;142(6):2876–88.
Houssein M, Abi Saab W, Khalil M, et al. Cell death by gallotannin is associated with inhibition of the JAK/STAT pathway in human colon cancer cells. Curr Ther Res Clin Exp. 2020;92: 100589.
Yang YL, Liu P, Li D, et al. Stat-3 signaling promotes cell proliferation and metastasis of gastric cancer through PDCD4 downregulation. Kaohsiung J Med Sci. 2020;36(4):244–9.
Zhang CS, Lin Y, Sun FB, Gao J, Han B, Li SJ. miR-409 down-regulates Jak-Stat pathway to inhibit progression of liver cancer. Eur Rev Med Pharmacol Sci. 2019;23(1):146–54.
Wang X, Wang J, Zhao J, Wang H, Chen J, Wu J. HMGA2 facilitates colorectal cancer progression via STAT3-mediated tumor-associated macrophage recruitment. Theranostics. 2022;12(2):963–75.
Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Bio-technol. 2017;35(4):314–6.
Wang X, Zhao Y, Dorothee FS, et al. The prognostic value of MicroRNAs associated with fatty acid metabolism in head and neck squamous cell carcinoma. Front Genet. 2022;30(13): 983672.
Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–88.
Zhao S, Zhang X, Gao F, et al. Identification of copper metabolism-related subtypes and establishment of prognostic model in ovarian cancer. Front Endocrinol. 2023;14:446.
Chi H, Jiang P, Ke X, et al. A novel anoikis-related gene signature predicts prognosis in patients with head and neck squamous cell carcinoma and reveals immune infiltration. Front Genet. 2022;26(13): 984273.
Yuan H, Yan M, Zhang G, et al. CancerSEA: a cancer single-cell state atlas. Nucl Acid Res. 2019;47:D900–8.
Shi J, Wei X, Xun Z, et al. The web-based portal SpatialTME integrates histological images with single-cell and spatial transcriptomics to explore the tumor microenvironment. Cancer Res. 2024;84(8):1210–20.
Xun Z, Ding X, Zhang Y, et al. Reconstruction of the tumor spatial microenvironment along the malignant-boundary-nonmalignant axis. Nat Commun. 2023;14:933.
Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013. https://doi.org/10.1126/scisignal.2004088.
Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50.
Julia CG, Eva S, Lisa T, et al. Microenvironment-induced CD44v6 promotes early disease progression in chronic lymphocytic leukemia. Blood. 2018;131(12):1337–49.
Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523.
Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable pro-tein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucl Acid Res. 2021;49(18):D605–12.
Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–2.
Li T, Fu J, Zeng Z, et al. TIMER20 for analysis of tumor-infiltrating immune cells. Nucl Acid Res. 2020. https://doi.org/10.1093/nar/gkaa407.
Chi H, Xie X, Yan Y, et al. Natural killer cell-related prognosis signature characterizes immune landscape and predicts prognosis of HNSCC. Front Immunol. 2022;13:1018685.
Zhao S, Chi H, Yang Q, et al. Identification and validation of neurotrophic factor-related gene signatures in glioblastoma and Parkinson’s disease. Front Immunol. 2023;14:1090040.
Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Sun D, Wang J, Han Y, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucl Acid Res. 2021;49:D1420–30.
Yan J, Ye G, Shao Y, et al. Identification of novel prognostic biomarkers in the TF-enhancer-target regulatory network in hepatocellular carcinoma and immune infiltration analysis. Front Genet. 2023;14:1158341.
Jin W, Yang Q, Chi H, et al. Ensemble deep learning enhanced with self-attention for predicting immunotherapeutic responses to cancers. Front Immunol. 2022;13:1025330.
Al-Danakh A, Safi M, Jian Y, et al. Aging-related biomarker discovery in the era of immune checkpoint inhibitors for cancer patients. Front Immunol. 2024;15:1348189.
Erdogan F, Qadree AK, Radu TB, et al. Structural and mutational analysis of mem-ber-specific STAT functions. Biochim Biophys Acta Gen Subj. 2021;1866(3): 130058.
Owen KL, Brockwell NK, Parker BS. JAK-STAT signaling: a double-edged sword of immune regulation and cancer pro-gression. Cancers (Basel). 2019;11(12):2002.
Gutiérrez-Hoya A, Soto-Cruz I. Role of the JAK/STAT pathway in cervical cancer: its relationship with HPV E6/E7 onco-proteins. Cells. 2020;9(10):2297.
Huang W, Li Y, Zhang C, et al. IGF2BP3 facilitates cell proliferation and tumorigenesis via modulation of JAK/STAT signalling pathway in human bladder cancer. J Cell Mol Med. 2020;24(23):13949–60.
Fang X, Hong Y, Dai L, et al. CRH promotes human colon cancer cell proliferation via IL-6/JAK2/STAT3 signaling pathway and VEGF-induced tumor angiogenesis. Mol Carcinog. 2017;56(11):2434–45.
She X, Xu J, Zhang H, et al. ETHE1 dampens colorectal cancer angiogenesis by promoting TC45 Dephosphorylation of STAT3 to inhibit VEGF-A expression. Cell Death Dis. 2024;15(8):631.
Du W, Wang YC, Hong J, et al. STAT5 isoforms regulate colorectal cancer cell apoptosis via reduction of mitochondrial membrane potential and generation of reactive oxygen species. J Cell Physiol. 2012;227(6):2421–9.
Keam SP, Halse H, Nguyen T, et al. High dose-rate brachytherapy of localized prostate cancer converts tumors from cold to hot. J Immunother Cancer. 2020;8(1): e000792.
Gong X, Chi H, Strohmer DF, et al. Exosomes: a potential tool for immunotherapy of ovarian cancer. Front Immunol. 2022;13:1089410.
Li J, Byrne KT, Yan F, et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 2018;49(1):178-193.e177.
Zhao Y, Wei K, Chi H, et al. IL-7: a promising adjuvant ensuring effective T cell responses and memory in combination with cancer vaccines? Front Immunol. 2022;13:1022808.
Lei X, Lei Y, Li JK, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–33.
Okuma A, Hanyu A, Watanabe S, et al. p16 and p21 promote tumour growth by enhancing myeloid-derived suppressor cells chemotaxis. Nat Commun. 2017;8(1):2050.
Kaneda MM, Cappello P, Nguyen AV, et al. Macrophage PI3Kγ drives pancreatic ductal adenocarcinoma progression. Cancer Discov. 2016;6(8):870–85.
Acknowledgements
We thank all contributors of high-quality data to these accessible public databases. We thank AJE for the language polishing work. This study was supported by grants from the Key Scientific and Technological Projects of Ningbo (No. 2021Z133), Ningbo Top Medical and Health Research Program (No.2023020612) and the Youth Medical Backbone Talents Training Program of Ningbo.
Funding
The Key Scientific and Technological Projects of Ningbo,2021Z133,the Youth Medical Backbone Talents Training Program of Ningbo, Ningbo Top Medical and Health Research Program,2023020612
Author information
Authors and Affiliations
Contributions
Yongfu Shao and Xuan Yu designed the study and critically reviewed the manuscript. Qier Li and Min Gan downloaded and analyzed the data. Qier Li and Qingqing Liu designed the figures. Min Gan and Jianing Yan wrote the manuscript. Yongfu Shao and Xuan Yu reviewed and edited the manuscript. Jingzhi Wang helped revised and polished the revised manuscript. The final manuscript has been approved by all authors.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Affiliated Hospital of Medical School of Ningbo University (No. KY20210762). All experiments were performed in accordance with relevant guidelines and regulations.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Q., Wang, J., Liu, Q. et al. Downregulated STAT3 and STAT5B are prognostic biomarkers for colorectal cancer and are associated with immune infiltration. Discov Onc 16, 343 (2025). https://doi.org/10.1007/s12672-025-02085-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-025-02085-2