Abstract
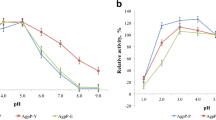
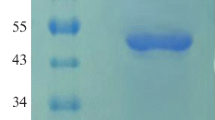
Phytases (myo-inositol hexakisphosphate hydrolase) catalyze the hydrolysis of phytate to inorganic phosphate and less phosphorylated myo-inositol derivatives and are widely used as feed additives in animal nutrition. Nevertheless, nowadays, there is a constant search for new phytases and new expression systems for better production of these important enzymes. In this study, we report cloning of two novel bacterial phytases belonging to the different enzyme classes and having different properties in the methylotrophic yeast Pichia pastoris. Sequences of agpP and phyC genes, encoding histidine acid phytase from Pantoea sp. 3.5.1 and β-propeller phytase from Bacillus ginsengihumi M2.11, respectively, were optimized and chemically synthesized according to the P. pastoris codon usage bias. The optimized genes were cloned into the yeast vectors pPINK-HC and pPINK-LC under the control of the inducible promoter AOX1 and two different signal peptides—signal sequence of α-amylase gene from Aspergillus niger and presequence of inulinase gene from Kluyveromyces maxianus. PCR analysis, restriction analysis, and DNA sequencing confirmed correct integration of agpP and phyC genes into P. pastoris genome. As a result, recombinant strains of P. pastoris with codon-optimized bacterial phytase genes (agpP and phyC), encoding histidine acid and β-propeller phytases, integrated into the genome under the alcohol oxidase promoter AOX1 and two different signal peptides, were obtained. Recombinant phytase AgpP was stably expressed and secreted into the culture medium of yeasts, whereas the expression of phyC gene was only confirmed at transcription level.






Similar content being viewed by others
References
Yao, M. Z., Zhang, Y. H., Lu, W. L., Hu, M. Q., Wang, W., & Liang, A. H. (2012). Phytases: crystal structures, protein engineering and potential biotechnological applications. J Appl Microbiol, 112, 1–14.
Maldonado, R. F., Maller, A., Bonneil, E., Thibault, P., Machado, C. B., Ward, R. J., et al. (2014). Biochemical properties of glycosylation and characterization of a histidine acid phosphatase (phytase) expressed in Pichia pastoris. Protein Expres Purif, 99, 43–49.
Olazaran, M. G., Blanco, L. R., Trevino, J. G. C., Lopez, J. A. G., & Salvado, J. M. V. (2010). Expression of a Bacillus phytase C gene in Pichia pastoris and properties of the recombinant enzyme. Applied and Environmental Microbiology, 76, 5601–5608.
Anchikov, E. V. (2012). Phytase in feeds for broilers J. Aviculture (Russian), 10, 22–25.
Ptak, A., Bedford, M. R., Świątkiewicz, S., Żyła, K., & Józefiak, D. (2015). Phytase modulates ileal microbiota and enhances growth performance of the broiler chickens. PLoS One, 10(3), e0119770.
Hurrell, J. W., Kushnir, Y., Ottersen, G., & Visbeck, M. (2003). The North Atlantic Oscillation. Climate Significance and Environmental Impact. Geophysical Monograph Series, 134.
Iqbal, T. H., Lewis, K. O., & Cooper, B. T. (1994). Phytase activity in the human and rat small intestine. Gut, 35, 1233–1236.
Sandberg, A. S., Hulthen, L. R., & Turk, M. (1996). Dietary Aspergillus niger phytase increases iron absorption in humans. The Journal of Nutrition, 126, 476–480.
Rao, K. V., Rao, T. P., & Reddy, V. D. (2009). Molecular characterization, physicochemical properties, known and potential applications of phytases: an overview. Critical Reviews in Biotechnology, 29(2), 182–198.
Siren, M. (1986a) Stabilized pharmaceutical and biological material composition. Patent SE 003165.
Siren, M. (1986b). New myo-inositol triphosphoric acid isomer. Patent SW, 052950.
Sariyska, M. V., Gargova, S. A., Koleva, L. A., & Angelov, A. I. (2005). Aspergillus niger phytase: purification and characterization. Biotechnology Equipment, 19, 98–105.
Wang, Y., Gao, X., Su, Q., Wu, W., & An, L. (2007). Expression of a heat stable phytase from Aspergillus fumigatus in tobacco (Nicotiana tabacum L. cv. NC89). Indian Journal of Biochemistry & Biophysics, 44, 26–30.
Zhang, L., An, L., Gao, X., & Wang, Y. (2004). Properties of A. ficuum AS3.324 phytase expressed in tobacco. Process Biochemistry, 40, 213–216.
Simon, O., & Igbasan, F. (2002). In vitro properties of phytases from various microbial origins. IJFST, 37, 813–822.
Quan, C., Fan, S., Zhang, L., Wang, Y., & Ohta, Y. (2002). Purfication and properties of a phytase from Candida krusei WZ-001. Journal of Bioscience and Bioengineering, 94, 419–425.
Hamada, A., Yamaguchi, K., Ohnishi, N., Harada, M., et al. (2005). High-level production of yeast (Schwanniomyces occidentalis) phytase in transgenic rice plants by a combination of signal sequence and codon modification of the phytase gene. Plant Biotechnology Journal, 3, 43–55.
Watanabe, T., Ikeda, H., Masaki, M., Fujii, T., & Iefuji, H. (2009). Cloning and characterization of a novel phytase from wastewater treatment yeast Hansenula fabianii J640 and expression in Pichia pastoris. Journal of Bioscience and Bioengineering, 108, 225–230.
Lei, X. G., Porres, J. M., Mullaney, E. J., Brinch, P. H. (2007). Industrial Enzyme: structure, function and applications. In: Springer Polaina J, MacCabe AP (ed.) . p. 505–529.
Menezes-Blackburn, D., Gabler, S., & Greiner, R. (2015). Performance of seven commercial phytases in an in vitro simulation of poultry digestive tract. Journal of Agricultural and Food Chemistry, 63, 6142–6149.
Adeola, L., & Zhai, H. (2011). Apparent and standardized ileal digestibilities of amino acids for pigs fed corn-soybean meal-based diets at varying crude protein levels. Journal of Animal Science, 89, 3626–3633.
Huang, H., Luo, H., Yang, P., Meng, K., Wang, Y., Yuan, T., et al. (2006). A novel phytase with preferable characteristics from Yersinia intermedia. Biochemical and Biophysical Research Communications, 350, 884–889.
Joshi, S., & Satyanarayana, T. (2014). Optimization of heterologous expression of the phytase (PPHY) of Pichia anomala in P. pastoris and its applicability in fractionating allergenic glycinin from soy protein. Journal of Industrial Microbiology & Biotechnology, 41, 977–987.
Kim, Y. O., Kim, H. W., Lee, J. H., Kim, K. K., & Lee, S. J. (2006). Molecular cloning of the phytase gene from Citrobacter braakii and its expression in Saccharomyces cerevisiae. Biotechnology Letters, 28, 33–38.
Promdonkoy, P., Tang, K., Sornlake, W., Harnpicharnchai, P., Kobayashi, R. S., Ruanglek, V., et al. (2009). Expression and characterization of Aspergillus thermostable phytases in Pichia pastoris. FEMS Microbiology Letters, 290, 18–24.
Daly, R., & Hearn, M. T. W. (2005). Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. Journal of Molecular Recognition, 18, 119–138.
Xiong, A., Yao, Q. H., Peng, R. H., Zhang, Z., Xu, F., Liu, J. G., et al. (2006). High level expression of a synthetic gene encoding Peniophora lycii phytase in methylotrphic yeast. Applied Microbiology and Biotechnology, 72, 1039–1047.
Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular cloning: a laboratory manual Cold Spring Harbor Laboratory press.
Cregg, J. M., & Russel, K. A. (1998). Transformation. Methods in Molecular Biology, 103, 27–39.
Schmitt, C. J., Zajicek, J. L., & Peterman, P. H. (1990). National contaminant biomonitoring program: residues of organochlorine chemicals in U.S. freshwater fish, 1976-1984. Archives of Environmental Contamination and Toxicology, 19, 748–781.
PichiaPink™ Expression System For high-level and large-scale expression and secretion of bioactive recombinant proteins in Pichia pastoris // Life Technologies.
Sambrook, J., Russell, D. (2001). Molecular cloning: A Laboratory manual (Third Edition). In: Chapter 4 working with high-capacity vectors. Cold Spring Harbor Laboratory Press. New York, USA.:42.
Greiner, R. (2004). Degradation of myo-inositol hexakisphosphate by a phytate-degrading enzyme from Pantoea agglomerans. The Protein J, 23, 577–585.
Suleimanova, A. D., Beinhauer, A., Valeeva, L. R., Chastukhina, I. B., Balaban, N. P., Shakirov, E. V., Greiner, R., & Sharipova, M. R. (2015). Novel glucose-1phosphatase with high phytase activity and unusual metal ion activation from soil bacterium Pantoea sp. strain 3.5.1. Applied and Environmental Microbiology, 81(19), 6790–6799.
Akbarzadeh, A., Ranaei Siadat, S. O., Motallebi, M., Zamani, M. R., Barshan Tashnizi, M., & Moshtaghi, S. (2014). Characterization and high level expression of acidic endoglucanase in Pichia pastoris. Applied Biochemistry and Biotechnology, 172(4), 2253–2265.
Bai, J., Swartz, D. J., Protasevich, I., Brouillette, C. G., et al. (2011). A gene optimization strategy that enhances production of fully functional P-glycoprotein in Pichia pastoris. PLoS One, 6(8), e22577.
Wang, J. R., Li, Y. Y., Liu, D. N., Liu, J. S., Li, P., Chen, L. Z. & Xu, SD (2015). Codon optimization significantly improves the expression level of α-amylase gene from Bacillus licheniformis in Pichia pastoris. BioMed Research International 2015. Article ID: 248680, 9 pages.
Obst, U., Lu, T. K., & Sieber, V. (2017). A modular toolkit for generating Pichia pastoris secretion. ACS Synthetic Biology, 6(6), 1016–1025.
Sreekrishna, K., Brankamp, R. G., Kropp, K. E., Blankenship, D. T., et al. (1997). Strategies for optimal synthesis and secretion of heterologous proteins in the methylotrophic yeast Pichia pastoris. Gene, 190, 55–62.
Cereghino, J. L., & Cregg, J. M. (2000). Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiology Reviews, 24(1), 45–66.
Gleeson, M. A., White, C. E., Meininger, D. P., & Komives, E. A. (1998). Generation of protease-deficient strains and their use in heterologous protein expression. Methods in Molecular Biology, 103, 81–94.
Csardi, G., Franks, A., Choi, D. S., Airoldi, E. M., Drummond, D. A. (2015). Accounting for experimental noise reveals that mRNA levels, amplified by post-transcriptional processes, largely determine steady-state protein levels in yeast. Snyder M, ed. PLoS Genetics, 11(5).
Greenbaum, D., Colangelo, C., Williams, K., & Gerstein, M. (2003). Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biology, 4(9), 117.
Acknowledgements
This work was performed in accordance with the Russian Government Program of Competitive Growth of Kazan Federal University.
Funding
This work was supported by the Russian Science Foundation (project no. 16-16-04062).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Troshagina, D.S., Suleimanova, A.D., Itkina, D.L. et al. Cloning of Phytase Genes from Pantoea Sp. 3.5.1 and Bacillus ginsengihumi M2.11 in Pichia pastoris. BioNanoSci. 8, 1045–1053 (2018). https://doi.org/10.1007/s12668-018-0563-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12668-018-0563-y