Abstract
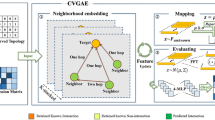
Long noncoding RNAs (lncRNAs) have significant regulatory roles in gene expression. Interactions with proteins are one of the ways lncRNAs play their roles. Since experiments to determine lncRNA–protein interactions (LPIs) are expensive and time-consuming, many computational methods for predicting LPIs have been proposed as alternatives. In the LPIs prediction problem, there commonly exists the imbalance in the distribution of positive and negative samples. However, there are few existing methods that give specific consideration to this problem. In this paper, we proposed a new clustering-based LPIs prediction method using segmented k-mer frequencies and multi-space clustering (LPI-SKMSC). It was dedicated to handling the imbalance of positive and negative samples. We constructed segmented k-mer frequencies to obtain global and local features of lncRNA and protein sequences. Then, the multi-space clustering was applied to LPI-SKMSC. The convolutional neural network (CNN)-based encoders were used to map different features of a sample to different spaces. It used multiple spaces to jointly constrain the classification of samples. Finally, the distances between the output features of the encoder and the cluster center in each space were calculated. The sum of distances in all spaces was compared with the cluster radius to predict the LPIs. We performed cross-validation on 3 public datasets and LPI-SKMSC showed the best performance compared to other existing methods. Experimental results showed that LPI-SKMSC could predict LPIs more effectively when faced with imbalanced positive and negative samples. In addition, we illustrated that our model was better at uncovering potential lncRNA–protein interaction pairs.
Graphic Abstract








Similar content being viewed by others
References
Mattick JS, Amaral PP, Carninci P et al (2023) Long non-coding rnas: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol 1–17. https://doi.org/10.1038/s41580-022-00566-8
Sun Y-M, Chen Y-Q (2020) Principles and innovative technologies for decrypting noncoding rnas: from discovery and functional prediction to clinical application. J Hematol Oncol 13:1–27. https://doi.org/10.1186/s13045-020-00945-8
Kung JT, Colognori D, Lee JT (2013) Long noncoding rnas: past, present, and future. Genetics 193(3):651–669. https://doi.org/10.1534/genetics.112.146704
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding rnas. RNA Biol 10(6):924–933. https://doi.org/10.4161/rna.24604
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding rnas: insights into functions. Nat Rev Genet 10(3):155–159. https://doi.org/10.1038/nrg2521
Nojima T, Proudfoot NJ (2022) Mechanisms of lncrna biogenesis as revealed by nascent transcriptomics. Nat Rev Mol Cell Biol 23(6):389–406. https://doi.org/10.1038/s41580-021-00447-6
Ravasi T, Suzuki H, Pang KC et al (2006) Experimental validation of the regulated expression of large numbers of non-coding rnas from the mouse genome. Genome Res 16(1):11–19. https://doi.org/10.1101/gr.4200206
Änkö M-L, Neugebauer KM (2012) Rna-protein interactions in vivo: global gets specific. Trends Biochem Sci 37(7):255–262. https://doi.org/10.1016/j.tibs.2012.02.005
Wang X, Arai S, Song X et al (2008) Induced ncrnas allosterically modify rna-binding proteins in cis to inhibit transcription. Nature 454(7200):126–130. https://doi.org/10.1038/nature06992
Wapinski O, Chang HY (2011) Long noncoding rnas and human disease. Trends Cell Biol 21(6):354–361. https://doi.org/10.1016/j.tcb.2011.04.001
Gao N, Li Y, Li J et al (2020) Long non-coding rnas: the regulatory mechanisms, research strategies, and future directions in cancers. Front Oncol 10:598817. https://doi.org/10.3389/fonc.2020.598817
Statello L, Guo C-J, Chen L-L et al (2021) Gene regulation by long non-coding rnas and its biological functions. Nat Rev Mol Cell Biol 22(2):96–118. https://doi.org/10.1038/s41580-020-00315-9
Esteller M (2011) Non-coding rnas in human disease. Nat Rev Genet 12(12):861–874. https://doi.org/10.1038/nrg3074
Sideris N, Dama P, Bayraktar S et al (2022) Lncrnas in breast cancer: A link to future approaches. Cancer Gene Ther., 1–12 https://doi.org/10.1038/s41417-022-00487-w
Simion V, Haemmig S, Feinberg MW (2019) Lncrnas in vascular biology and disease. Vascul Pharmacol 114:145–156. https://doi.org/10.1016/j.vph.2018.01.003
Aznaourova M, Schmerer N, Schmeck B et al (2020) Disease-causing mutations and rearrangements in long non-coding rna gene loci. Front Genet 11:527484. https://doi.org/10.3389/fgene.2020.527484
Ray D, Kazan H, Chan ET et al (2009) Rapid and systematic analysis of the rna recognition specificities of rna-binding proteins. Nat Biotechnol 27(7):667–670. https://doi.org/10.1038/nbt.1550
Keene JD, Komisarow JM, Friedersdorf MB (2006) Rip-chip: the isolation and identification of mrnas, micrornas and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1(1):302–307. https://doi.org/10.1038/nprot.2006.47
Licatalosi DD, Mele A, Fak JJ et al (2008) Hits-clip yields genome-wide insights into brain alternative rna processing. Nature 456(7221):464–469. https://doi.org/10.1038/nature07488
Hafner M, Landthaler M, Burger L et al (2010) Transcriptome-wide identification of rna-binding protein and microrna target sites by par-clip. Cell 141(1):129–141. https://doi.org/10.1016/j.cell.2010.03.009
Bellucci M, Agostini F, Masin M et al (2011) Predicting protein associations with long noncoding rnas. Nat Methods 8(6):444–445. https://doi.org/10.1038/nmeth.1611
Muppirala UK, Honavar VG, Dobbs D (2011) Predicting rna-protein interactions using only sequence information. BMC Bioinform. 12(1):1–11. https://doi.org/10.1186/1471-2105-12-489
Lu Q, Ren S, Lu M et al (2013) Computational prediction of associations between long non-coding rnas and proteins. BMC Genom 14(1):1–10. https://doi.org/10.1186/1471-2164-14-651
Suresh V, Liu L, Adjeroh D et al (2015) Rpi-pred: predicting ncrna-protein interaction using sequence and structural information. Nucleic Acids Res 43(3):1370–1379. https://doi.org/10.1093/nar/gkv020
Li A, Ge M, Zhang Y et al (2015) Predicting long noncoding rna and protein interactions using heterogeneous network model. BioMed Res Int 2015. https://doi.org/10.1155/2015/671950
Ge M, Li A, Wang M (2016) A bipartite network-based method for prediction of long non-coding rna-protein interactions. Genom Proteom Bioinf 14(1):62–71. https://doi.org/10.1016/j.gpb.2016.01.004
Pan X, Fan Y-X, Yan J et al (2016) Ipminer: hidden ncrna-protein interaction sequential pattern mining with stacked autoencoder for accurate computational prediction. BMC Genom 17:1–14. https://doi.org/10.1186/s12864-016-2931-8
Wekesa JS, Luan Y, Chen M et al (2019) A hybrid prediction method for plant lncrna-protein interaction. Cells 8(6):521. https://doi.org/10.3390/cells8060521
Zhang S-W, Zhang X-X, Fan X-N et al (2020) Lpi-cnncp: prediction of lncrna-protein interactions by using convolutional neural network with the copy-padding trick. Anal Biochem 601:113767. https://doi.org/10.1016/j.ab.2020.113767
Huang L, Jiao S, Yang S et al (2021) Lgfc-cnn: prediction of lncrna-protein interactions by using multiple types of features through deep learning. Genes 12(11):1689. https://doi.org/10.3390/genes12111689
Shen Z-A, Luo T, Zhou Y-K et al (2021) Npi-gnn: Predicting ncrna-protein interactions with deep graph neural networks. Brief. Bioinformatics 22(5):051. https://doi.org/10.1093/bib/bbab051
Fan X-N, Zhang S-W (2019) Lpi-bls: Predicting lncrna-protein interactions with a broad learning system-based stacked ensemble classifier. Neurocomputing 370:88–93. https://doi.org/10.1016/j.neucom.2019.08.084
Zhou H, Wekesa JS, Luan Y et al (2021) Prpi-sc: an ensemble deep learning model for predicting plant lncrna-protein interactions. BMC Bioinform. 22(3):1–15. https://doi.org/10.1186/s12859-021-04328-9
Zhuo L, Song B, Liu Y et al (2022) Predicting ncrna-protein interactions based on dual graph convolutional network and pairwise learning. Brief. Bioinformatics 23(6):339. https://doi.org/10.1093/bib/bbac339
Tian X, Shen L, Wang Z et al (2021) A novel lncrna-protein interaction prediction method based on deep forest with cascade forest structure. Sci Rep 11(1):18881. https://doi.org/10.1038/s41598-021-98277-1
Lihong P, Wang C, Tian X et al (2021) Finding lncrna-protein interactions based on deep learning with dual-net neural architecture. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2021.3116232
Song J, Tian S, Yu L et al (2022) Rlf-lpi: An ensemble learning framework using sequence information for predicting lncrna-protein interaction based on ae-reslstm and fuzzy decision. Math Biosci Eng 19(5):4749–4764. https://doi.org/10.3934/mbe.2022222
Zhou L, Wang Z, Tian X et al (2021) Lpi-deepgbdt: a multiple-layer deep framework based on gradient boosting decision trees for lncrna-protein interaction identification. BMC Bioinform. 22(1):1–24. https://doi.org/10.1186/s12859-021-04399-8
Yu B, Wang X, Zhang Y et al (2022) Rpi-mdlstack: Predicting rna-protein interactions through deep learning with stacking strategy and lasso. Appl Soft Comput 120:108676. https://doi.org/10.1016/j.asoc.2022.108676
Hao Y, Wu W, Li H et al (2016) Npinter v3. 0: an upgraded database of noncoding rna-associated interactions. Database 2016https://doi.org/10.1093/database/baw057
Apweiler R, Bairoch A, Wu CH et al (2004) Uniprot: the universal protein knowledgebase. Nucleic Acids Res. 32(suppl_1):115–119 https://doi.org/10.1093/nar/gkh131
Frankish A, Diekhans M, Ferreira A-M et al (2019) Gencode reference annotation for the human and mouse genomes. Nucleic Acids Res 47(D1):766–773. https://doi.org/10.1093/nar/gky955
Berman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Lewis BA, Walia RR, Terribilini M et al (2010) Pridb: a protein–rna interface database. Nucleic Acids Res. 39(suppl_1):277–282 https://doi.org/10.1093/nar/gkq1108
Ruff L, Vandermeulen RA, Görnitz N et al (2019) Deep semi-supervised anomaly detection. arXiv preprint arXiv:1906.02694
Ruff L, Vandermeulen R, Goernitz N et al (2018) Deep one-class classification, 4393–4402. PMLR. https://proceedings.mlr.press/v80/ruff18a.html
Wang L, Wang H-F, Liu S-R et al (2019) Predicting protein-protein interactions from matrix-based protein sequence using convolution neural network and feature-selective rotation forest. Sci Rep 9(1):9848. https://doi.org/10.1038/s41598-019-46369-4
Soleymani F, Paquet E, Viktor HL et al (2023) Protinteract: A deep learning framework for predicting protein–protein interactions. Comput Struct Biotechnol J 21:1324–1348. https://doi.org/10.1016/j.csbj.2023.01.028
Yang X, Yang S, Lian X et al (2021) Transfer learning via multi-scale convolutional neural layers for human-virus protein-protein interaction prediction. Bioinformatics 37(24):4771–4778. https://doi.org/10.1093/bioinformatics/btab533
Liu K, Cao L, Du P et al (2020) im6a-ts-cnn: identifying the n6-methyladenine site in multiple tissues by using the convolutional neural network. Mol. Ther. Nucleic. Acids 21:1044–1049. https://doi.org/10.1016/j.omtn.2020.07.034
Alam W, Ali SD, Tayara H et al (2020) A cnn-based rna n6-methyladenosine site predictor for multiple species using heterogeneous features representation. IEEE Access 8:138203–138209. https://doi.org/10.1109/ACCESS.2020.3002995
Zhang P, Meng J, Luan Y et al (2020) Plant mirna–lncrna interaction prediction with the ensemble of cnn and indrnn. Interdiscip Sci 12:82–89. https://doi.org/10.1007/s12539-019-00351-w
Kang Q, Meng J, Cui J et al (2020) Pmlipred: a method based on hybrid model and fuzzy decision for plant mirna–lncrna interaction prediction. Bioinformatics 36(10):2986–2992. https://doi.org/10.1093/bioinformatics/btaa074
Yang S, Wang Y, Lin Y et al (2020) Lncmirnet: predicting lncrna-mirna interaction based on deep learning of ribonucleic acid sequences. Molecules 25(19):4372. https://doi.org/10.3390/molecules25194372
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Data availability
The source code and datasets for this study are accessible at https://github.com/zhlSunLab/LPI-SKMSC.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, DZ., Sun, ZL., Liu, M. et al. LPI-SKMSC: Predicting LncRNA–Protein Interactions with Segmented k-mer Frequencies and Multi-space Clustering. Interdiscip Sci Comput Life Sci (2024). https://doi.org/10.1007/s12539-023-00598-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12539-023-00598-4