Abstract
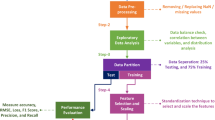
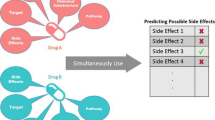
Hypertension (HT) is a general disease, and also one of the most ordinary and major causes of cardiovascular disease. Some diseases are caused by high blood pressure, including impairment of heart and kidney function, cerebral hemorrhage and myocardial infarction. Due to the limitations of laboratory methods, bioactive peptides for the treatment of HT need a long time to be identified. Therefore, it is of great immediate significance for the identification of anti-hypertensive peptides (AHTPs). With the prevalence of machine learning, it is suggested to use it as a supplementary method for AHTPs classification. Therefore, we develop a new model to identify AHTPs based on multiple features and deep learning. And the deep model is constructed by combining a convolutional neural network (CNN) and a gated recurrent unit (GRU). The unique convolution structure is used to reduce the feature dimension and running time. The data processed by CNN is input into the recurrent structure GRU, and important information is filtered out through the reset gate and update gate. Finally, the output layer adopts Sigmoid activation function. Firstly, we use Kmer, the deviation between the dipeptide frequency and the expected mean (DDE), encoding based on grouped weight (EBGW), enhanced grouped amino acid composition (EGAAC) and dipeptide binary profile and frequency (DBPF) to extract features. For Kmer, DDE, EBGW and EGAAC, it is widely used in the field of protein research. DBPF is a new feature representation method designed by us. It corresponds dipeptides to binary numbers, and finally obtains a binary coding file and a frequency file. Then these features are spliced together and input into our proposed model for prediction and analysis. After a tenfold cross-validation test, this model has a better competitive advantage than the previous methods, and the accuracy is 96.23% and 99.10%, respectively. From the results, compared with the previous methods, it has been greatly improved. It shows that the combination of convolution calculation and recurrent structure has a positive impact on the classification of AHTPs. The results show that this method is a feasible, efficient and competitive sequence analysis tool for AHTPs. Meanwhile, we design a friendly online prediction tool and it is freely accessible at http://ahtps.zhanglab.site/.
Graphical Abstract








Similar content being viewed by others
References
Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Rohani HA et al (2012) A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380(9859):2224–2260. https://doi.org/10.1016/S0140-6736(12)61766-8
Chockalingam A, Campbell NR, Fodor JG (2006) Worldwide epidemic of hypertension. Can J Cardiol 22(7):553–555. https://doi.org/10.1016/s0828-282x(06)70275-6
Thomopoulos C, Parati G, Zanchetti A (2014) Effects of blood pressure lowering on outcome incidence in hypertension: 2. Effects at different baseline and achieved blood pressure levels-overview and meta-analyses of randomized trials. J Hypertens 32(12):2296–2304. https://doi.org/10.1097/HJH.0000000000000379
Varounis C, Katsi V, Nihoyannopoulos P, Lekakis J, Tousoulis D (2016) Cardiovascular hypertensive crisis: recent evidence and review of the literature. Front Cardiovasc Med 3:51. https://doi.org/10.3389/fcvm.2016.00051
Husserl FE, Messerli FH (1981) Adverse effects of antihypertensive drugs. Drugs 22(3):188–210. https://doi.org/10.2165/00003495-198122030-00002
Ledesma BH, Contreras MDM, Recio I (2011) Antihypertensive peptides: production, bioavailability and incorporation into foods. Adv Colloid Interface 165(1):23–35. https://doi.org/10.1016/j.cis.2010.11.001
Saito T (2008) Antihypertensive peptides derived from bovine casein and whey proteins. Adv Exp Med Biol 606:295–317. https://doi.org/10.1007/978-0-387-74087-4_12
Escudero E, Toldrá F, Sentandreu MA, Nishimura H, Arihara K (2012) Antihypertensive activity of peptides identified in the in vitro gastrointestinal digest of pork meat. Meat Sci 91(3):382–384. https://doi.org/10.1016/j.meatsci.2012.02.007
Kitts DD, Weiler K (2003) Bioactive proteins and peptides from food sources. Applications of bioprocesses used in isolation and recovery. Curr Pharm Des 9(16):1309–1323. https://doi.org/10.2174/1381612033454883
Dostal DE, Baker KM (1999) The cardiac renin-angiotensin system: conceptual, or a regulator of cardiac function? Circ Res 85(7):643–650. https://doi.org/10.1161/01.res.85.7.643
Bhat ZF, Kumar S, Bhat HF (2017) Antihypertensive peptides of animal origin: a review. Crit Rev Food Sci 57(3):566–578. https://doi.org/10.1080/10408398.2014.898241
Jakala P, Vapaatalo H (2010) Antihypertensive peptides from milk proteins. Pharmaceuticals 3(1):251–272. https://doi.org/10.3390/ph3010251
Majumder K, Wu J (2014) Molecular targets of antihypertensive peptides: understanding the mechanisms of action based on the pathophysiology of hypertension. Int J Mol Sci 16(1):256–283. https://doi.org/10.3390/ijms16010256
Puchalska P, Alegre MLM, López MCG (2015) Isolation and characterizeati-on of peptides with antihypertensive activity in foodstuffs. Crit Rev Food Sci 55:521–551. https://doi.org/10.1080/10408398.2012.664829
Wang XY, Wang J, Lin Y, Ding Y, Wang YQ, Cheng XM, Lin ZH (2011) QSAR study on angiotensin-converting enzyme inhibitor oligopeptides based on a novel set of sequence information descriptors. J Mol Model 17:1599–1606. https://doi.org/10.1007/s00894-010-0862-x
Kumar R, Chaudhary K, Chauhan JS, Nagpal G, Kumar R, Sharma M, Raghava GPS (2015) An in silico platform for predicting, screening and designing of antihypertensive peptides. Sci Rep 5:12512. https://doi.org/10.1038/srep12512
Win TS, Schaduangrat N, Prachayasittikul V, Nantasenamat C, Shoombuato-ng W (2018) PAAP: a web server for predicting antihypertensive activity of peptides. Fut Med Chem 10(15):1749–1767. https://doi.org/10.4155/fmc-2017-0300
Manavalan B, Basith S, Shin TH, Wei LY, Lee G (2019) mAHTPred: a sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics 35(16):2757–2765. https://doi.org/10.1093/bioinformatics/bty1047
Zhuang YY, Liu XR, Zhong Y, Wu LX (2021) A deep ensemble predictor for identifying anti-hypertensive peptides using pretrained protein embedding. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2021.3068381
Alley EC, Khimulya G, Biswas S, AlQuraishi M, Church GM (2019) Unified rational protein engineering with sequence-based deep representation learning. Nat Methods 16:1315–1322. https://doi.org/10.1038/s41592-019-0598-1
Öztürk Ş (2020) Two-stage sequential losses based automatic hash code generation using Siamese network. Avrupa Bilim ve Teknoloji Dergisi. https://doi.org/10.31590/ejosat.801927
Öztürk Ş (2021) Convolutional neural network based dictionary learning to create hash codes for content-based image retrieval. Proc Comput Sci 183:624–629. https://doi.org/10.1016/j.procs.2021.02.106
Öztürk Ş, Alhudhaif A, Polat K (2021) Attention-based end-to-end CNN framework for content-based X-ray image retrieval. Turk J Electr Eng Comput Sci 29:2680–2693. https://doi.org/10.3906/elk-2105-242
Chung J, Gulcehre C, Cho K, Bengio Y (2014) Empirical evaluation of gated recurrent neural networks on sequence modeling. Neural Evol Comput. https://doi.org/10.48550/arXiv.1412.3555
Yi YH, Lv YY, Zhang LJ, Yang J, Shi Q (2018) High throughput identification of antihypertensive peptides from fish proteome datasets. Mar Drugs 16(10):365. https://doi.org/10.3390/md16100365
Kumar R, Chaudhary K, Sharma M, Nagpal G, Chauhan JS, Singh S, Gautam A, Raghava GPS (2015) AHTPDB: a comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Res 43(D1):D956–D962. https://doi.org/10.1093/nar/gku1141
Iwaniak A, Minkiewicz P, Darewicz M, Sieniawski K, Starowicz P (2016) BIOPEP database of sensory peptides and amino acids. Food Res Int 85:155–161. https://doi.org/10.1016/j.foodres.2016.04.031
Agrawal P, Bhalla S, Chaudhary K, Kumar R, Sharma M, Raghava GPS (2018) In silico approach for prediction of antifungal peptides. Front Microbiol 9:323. https://doi.org/10.3389/fmicb.2018.00323
Chen W, Ding H, Feng PM, Lin H (2016) iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget 7(13):16895–16909. https://doi.org/10.18632/oncotarget.7815
Sharma A, Kapoor P, Gautam A, Chaudhary K, Kumar R, Chauhan JS, Tyagi A, Raghava GPS (2013) Computational approach for designing tumor homing peptides. Sci Rep 3:1607. https://doi.org/10.1038/srep01607
Usmani SS, Bhalla S, Raghava GPS (2018) Prediction of antitubercular peptides from sequence information using ensemble classifier and hybrid features. Front Pharmacol 9:954. https://doi.org/10.3389/fphar.2018.00954
Wei L, Zhou C, Chen HR, Song JN, Su R (2018) ACPred-FL: a sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics 34(23):4007–4016. https://doi.org/10.1093/bioinformatics/bty451
Saravanan V, Gautham N (2015) Harnessing computational biology for exact linear B-cell epitope prediction: a novel amino acid composition-based feature descriptor. OMICS 19(10):648–658. https://doi.org/10.1089/omi.2015.0095
Zhang ZH, Wang ZH, Zhang ZR, Wang YX (2006) A novel method for apoptosis protein subcellular localization prediction combining encoding based on grouped weight and support vector machine. FEBS Lett 580(26):6169–6174. https://doi.org/10.1016/j.febslet.2006.10.017
Wang XY, Yu B, Ma AJ, Chen C, Liu BQ, Ma Q (2019) Protein-protein interaction sites prediction by ensemble random forests with synthetic minority oversampling technique. Bioinformatics 35(14):2395–2402. https://doi.org/10.1093/bioinformatics/bty995
Tian BG, Wu X, Chen C, Qiu WY, Ma Q, Yu B (2019) Predicting protein-protein interactions by fusing various Chou’s pseudo components and using wavelet denoising approach. J Theor Biol 462:329–346. https://doi.org/10.1016/j.jtbi.2018.11.011
Yu B, Qiu WY, Chen C, Ma AJ, Jiang J, Zhou HY, Ma Q (2019) SubMito-XGBoost: predicting protein submitochondrial localization by fusing multiple feature information and eXtreme gradient boosting. Bioinformatics 36(4):1074–1081. https://doi.org/10.1093/bioinformatics/btz734
Yu B, Yu ZM, Chen C, Ma AJ, Liu BQ, Tian BG, Ma Q (2020) DNNAce: prediction of prokaryote lysine acetylation sites through deep neural networks with multi-information fusion. Chemometr Intell Lab 200:103999. https://doi.org/10.1016/j.chemolab.2020.103999
Lee TY, Lin ZQ, Hsieh SJ, Bretaña NA, Lu CT (2011) Exploiting maximal dependence decomposition to identify conserved motifs from a group of aligned signal sequences. Bioinformatics 27(13):1780–1787. https://doi.org/10.1093/bioinformatics/btr291
Li Y, Zheng WM, Cui Z, Zhang T (2018) Face recognition based on recurrent regression neural network. Neurocomputing 297:50–58. https://doi.org/10.1016/j.neucom.2018.02.037
Morchid M (2018) Parsimonious memory unit for recurrent neural networks with application to natural language processing. Neurocomputing 2018(314):48–64. https://doi.org/10.1016/j.neucom.2018.05.081
Connor JT, Martin RD, Atlas LE (1994) Recurrent neural networks and robust time series prediction. IEEE T Neural Networ 5(2):240–254. https://doi.org/10.1109/72.279188
Nguyen QKL (2019) Fertility-GRU: identifying fertility-related proteins by incorporating deep-gated recurrent units and original position-specific scoring matrix profiles. J Proteome Res 18(9):3503–3511. https://doi.org/10.1021/acs.jproteome.9b00411
Li YW, Golding GB, Ilie L (2021) DELPHI: accurate deep ensemble model for protein interaction sites prediction. Bioinformatics 37(7):896–904. https://doi.org/10.1093/bioinformatics/btaa750
Wang MH, Cui XW, Li S, Yang XH, Ma AJ, Zhang YS, Yu B (2020) DeepMal: Accurate prediction of protein malonylation sites by deep neural networks. Chemometr Intell Lab 207:104175. https://doi.org/10.1016/j.chemolab.2020.104175
Liang JX, Cui ZF, Wu CB, Yu Y, Tian R, Xie HX, Jin Z, Fan WW, Xie WL, Huang ZY, Xu W, Zhu JJ, You ZS, Guo XF, Qiu XF, Ye JH, Lang B, Li MY, Tan SW, Hu Z (2021) DeepEBV: a deep learning model to predict Epstein-Barr virus (EBV) integration sites. Bioinformatics. https://doi.org/10.1093/bioinformatics/btab388
Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):1929–1958. https://doi.org/10.5555/2627435.2670313
Huo YH, Xin LH, Kang CZ, Wang MH, Ma Q, Yu B (2020) SGL-SVM: A novel method for tumor classification via support vector machine with sparse group Lasso. J Theor Biol 486:110098. https://doi.org/10.1016/j.jtbi.2019.110098
Chen C, Zhang QM, Ma Q, Yu B (2019) LightGBM-PPI: Predicting protein-protein interactions through LightGBM with multi-information fusion. Chemometr Intell Lab 191:54–64. https://doi.org/10.1016/j.chemolab.2019.06.003
Shi H, Liu SM, Chen JQ, Li X, Ma Q, Yu B (2019) Predicting drug-target interactions using Lasso with random forest based on evolutionary information and chemical structure. Genomics 111(6):1839–1852. https://doi.org/10.1016/j.ygeno.2018.12.007
Chen C, Zhang QM, Yu B, Yu ZM, Lawrence PJ, Ma Q, Zhang Y (2020) Improving protein–protein interactions prediction accuracy using XGBoost feature selection and stacked ensemble classifier. Comput Biol Med 123:103899. https://doi.org/10.1016/j.compbiomed.2020.103899
Sun XM, Jin TY, Chen C, Cui XW, Ma Q, Yu B (2020) RBPro-RF: Use Chou’s 5-steps rule to predict RNA-binding proteins via random forest with elastic net. Chemometr Intell Lab 197:103919. https://doi.org/10.1016/j.chemolab.2019.103919
Zhang Q, Li S, Yu B, Zhang QM, Han Y, Zhang Y, Ma Q (2020) DMLDA-LocLIFT: identification of multi-label protein subcellular localizati-on using DMLDA dimensionality reduction and LIFT classifier. Chemometr Intell Lab Syst 206:104148. https://doi.org/10.1016/j.chemolab.2020.104148
Zhang SL, Qiao HJ (2020) KD-KLNMF: identification of lncRNAs subcellular localization with multiple features and nonnegative matrix factorization. Anal Biochem 610:113995. https://doi.org/10.1016/j.ab.2020.113995
Zhang SL, Xue T (2020) Use Chous 5-steps rule to identify DNase I hypersensitive sites via dinucleotide property matrix and extreme gradient boosting. Mol Genet Genomics 295:1431–1442. https://doi.org/10.1007/s00438-020-01711-8
Zhang YP, Zou Q (2020) PPTPP: a novel therapeutic peptide prediction method using physicochemical property encoding and adaptive feature representation learning. Bioinformatics 36(13):3982–3987. https://doi.org/10.1093/bioinformatics/btaa275
Wang JY, Zhang SL, Qiao HJ, Wang JS (2021) UMAP-DBP: an improved DNA-binding proteins prediction method based on uniform manifold approximation and projection. Protein J 40(4):562–575. https://doi.org/10.1007/s10930-021-10011-y
Wei LY, Luan SS, Nagai LAE, Su R, Zou Q (2019) Exploring sequence-based features for the improved prediction of DNA N4-methylcytosine sites in multiple species. Bioinformatics 35(8):1326–1333. https://doi.org/10.1093/bioinformatics/bty824
Zou Q, Xing PW, Wei LY, Liu B (2019) Gene2vec: gene subsequence embedding for prediction of mammalian N6-methyladenosine sites from mRNA. RNA 25(2):205–218. https://doi.org/10.1261/rna.069112.118
Zhao ZX, Zhang XC, Chen F, Fang L, Li JY (2020) Accurate prediction of DNA N4-methylcytosine sites via boost-learning various types of sequence features. BMC Genomics 21(1):627. https://doi.org/10.1186/s12864-020-07033-8
Wang JS, Zhang SL (2021) PA-PseU: an incremental passive-aggressive based method for identifying RNA pseudouridine sites via Chou’s 5-steps rule. Chemometr Intell Lab 210:104250. https://doi.org/10.1016/j.chemolab.2021.104250
Singh VK, Maurya NS, Mani A, Yadav RS (2020) Machine learning method using position-specific mutation based classification outperforms one hot coding for disease severity prediction in haemophilia ‘A.’ Genomics 112(6):5122–5128. https://doi.org/10.1016/j.ygeno.2020.09.020
Xie YB, Luo XT, Li YP, Chen L, Ma WB, Huang JJ, Cui J, Zhao Y, Xue Y, Zuo ZX, Ren J (2018) DeepNitro: prediction of protein nitration and nitrosylation sites by deep learning. Genom Proteom Bioinf 16(4):294–306. https://doi.org/10.1016/j.gpb.2018.04.007
Liu Q, Xia F, Yin QJ, Jiang R (2018) Chromatin accessibility prediction via a hybrid deep convolutional neural network. Bioinformatics 34(5):732–738. https://doi.org/10.1093/bioinformatics/btx679
Hamid MN, Friedberg I (2019) Identifying antimicrobial peptides using word embedding with deep recurrent neural networks. Bioinformatics 35(12):2009–2016. https://doi.org/10.1093/bioinformatics/bty937
Chou KC (2015) Impacts of bioinformatics to medicinal chemistry. Med Chem 11(3):218–234. https://doi.org/10.2174/1573406411666141229162834
Xue T, Zhang SL, Qiao HJ (2021) i6mA-VC: a multi-classifier voting method for the computational identification of DNA N6-methyladenine sites. Interdiscip Sci 13(3):413–425. https://doi.org/10.1007/s12539-021-00429-4
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No.12101480), the Natural Science Basic Research Program of Shaanxi (No.2021JM-115), and the Fundamental Research Funds for the Central Universities (No. JB210715).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Shi, H., Zhang, S. Accurate Prediction of Anti-hypertensive Peptides Based on Convolutional Neural Network and Gated Recurrent unit. Interdiscip Sci Comput Life Sci 14, 879–894 (2022). https://doi.org/10.1007/s12539-022-00521-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-022-00521-3