Abstract
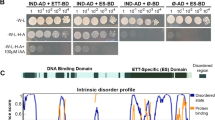
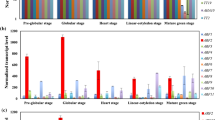
Some Arabidopsis thaliana AUXIN/INDOLE ACETIC ACIDs (Aux/IAAs) were predicted to have two ETHYLENE RESPONSE FACTOR–associated Amphiphilic Repression (EAR) motifs which would be involved in repression of auxin responses by interacting with TOPLESS (TPL)/TPL-Related (TPR) corepressors. However, the function of the 2nd EAR motif and any distinctive functions of two EAR motifs have remained to be characterized. Here, we analyzed the biological and molecular function of two EAR motifs of IAA7 by using the substitution mutant forms of the EAR motifs. The 2nd EAR motif played a minor (compared with the 1st one) repressive role in auxin-related developmental processes such as root hair growth, primary root growth, cotyledon curling, and plant dwarfism, and also in auxinresponsive gene expression. The yeast two-hybrid assay revealed that while the 1st EAR motif played a dominant role in interacting with all TPL/TPR members, the 2nd EAR was specifically required to interact with TPR1 and played a minor role in interaction with TPL. The protein pull-down analysis further supported the engagement of the 2nd EAR motif in interaction with TPL and TPR1. These results indicate that two EAR motifs of IAA7 plays repressive roles in auxin responses by interacting with TPL/TPRs with specificity.
Similar content being viewed by others
References
Calderón-Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, Zheng N, Napier R, Kepinski S, Estelle M (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8:477–485
Causier B, Ashworth M, Guo W, Davies B (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158:423–438
Cho H-T, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14:3237–3253
Cho M, Lee SH, Cho H-T (2007) P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19:3930–3943
Dharmasiri N, Dharmasiri S, Estelle M (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445
Dharmasiri N, Dharmasiri S,Weijers D, Lechner E, YamadaM, Hobbie L, Ehrismann J, Jürgens G, Estelle M (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9:109–119
Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18:699–714
Ganguly A, Lee SH, Cho M, Lee OR, Ryu H, Cho H-T (2010) Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol 153:1046–1061
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414:271–276
Guilfoyle TJ (2015) The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27:33–43
Guilfoyle TJ, Hagen G (2012) Getting a grasp on domain III/IV responsible for Auxin Response Factor–IAA protein interactions. Plant Sci 190:82–88
Hagen G, Guilfoyle TJ (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385
Han M, Park Y, Kim I, Kim E-H, Yu TK, Rhee S, Suh J-Y (2014) Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc Natl Acad Sci USA 111:18613–18618
Kagale S, Links MG, Rozwadowski K (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152:1109–1134
Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435:446–451
Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho H-T (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18:2958–2970
Knox K, Grierson CS, Leyser O (2003) AXR3 and SHY2 interact to regulate root hair development. Development 130:5769–5777
Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC (2014) Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA 111:5427–5432
Lee M-S, Choi H-S, Cho H-T (2013) Branching the auxin signaling; multiple players and diverse interactions. J Plant Biol 56:130–137
Lee SH, Cho H-T (2006) PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18:1604–1616
Lee SH, Cho H-T (2009) Auxin and Root Hair Morphogenesis, In AM Emons, T Ketelaar, eds, Root Hairs, Ed 1, Vol 12. Springer-Verlag Berlin Heideberg, pp 45–64
Leyser HM, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10:403–413
Li J-F, Bush J, Xiong Y, Li L, McCormack M (2011a) Large-scale protein-protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PLoS ONE 6:e27364
Li H, Cheng Y, Murphy A, Hagen G, Guilfoyle TJ (2009) Constitutive repression and activation of auxin signaling in Arabidopsis. Plant Physiol 149: 1277–1288
Li H, Hagen G, Guilfoyle TJ (2011b) Do some IAA proteins have two repression domains? Plant Signal Behav 6:858–860
Li H, Tiwari SB, Hagen G, Guilfoyle TJ (2011c) Identical amino acid substitutions in the repression domain of auxin/indole-3-acetic acid proteins have contrasting effects on auxin signaling. Plant Physiol 155:1252–1263
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49:387–400
Lokerse AS, Weijers D (2009) Auxin enters the matrix-assembly of response machineries for specific outputs. Cur Opi Plant Biol 12:520–526
Masucci JD, Schiefelbein JW (1994) The rhd6 mutation of Arabidopsis thaliana alters root hair initiation through an auxin-and ethylene-associated process. Plant Physiol 106:1335–1346
Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24:55–80
Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT (2007) Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoterexchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol 144:187–196
Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123:563–574
Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25:213–221
Pérez-Pérez JM, Candela H, Robles P, López-Torrejón G, del Pozo JC, Micol JL (2010) A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol 51:1661–1673
Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6:420–425
Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135:1738–1752
Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27:9–19.
Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384–1386
Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–645
Tiwari SB, Hagen G, Guilfoyle TJ (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543
Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16:533–543
Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13:2809–2822
Vernoux T, Brunoud G, Farcot E, Morin V, den Daele HV, Legrand J, Oliva M, Das P, Larrieu A, Wells D, Guedon Y, Armitage L, Picard F, Guyomarc’h S, Cellier C, Parry G, Koumproglou R, Doonan JH, Estelle M, Godin C, Kepinski S, Bennett M, De Veylder L, Traas J (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Sys Biol 7:508522
Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Klentz M, Wilmoth JC, Reed JW, Jürgens G (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24:1874–1885
Wilson AK, Pickett FB, Turner JC, Estelle M (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222:377–383
Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho H-T (2009) Ciselement-and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol 150:1459–1473
Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21:553–562
Yi K, Menand B, Bell E, Dolan L (2010) A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Gen 42:264–267
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lee, MS., An, JH. & Cho, HT. Biological and molecular functions of two EAR motifs of Arabidopsis IAA7. J. Plant Biol. 59, 24–32 (2016). https://doi.org/10.1007/s12374-016-0453-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-016-0453-1