Abstract
Currently, salinization is impacting more than 50% of arable land, posing a significant challenge to agriculture globally. Salt causes osmotic and ionic stress, determining cell dehydration, ion homeostasis, and metabolic process alteration, thus negatively influencing plant development. A promising sustainable approach to improve plant tolerance to salinity is the use of plant growth-promoting bacteria (PGPB). This work aimed to characterize two bacterial strains, that have been isolated from pea root nodules, initially called PG1 and PG2, and assess their impact on growth, physiological, biochemical, and molecular parameters in three pea genotypes (Merveille de Kelvedon, Lincoln, Meraviglia d’Italia) under salinity. Bacterial strains were molecularly identified, and characterized by in vitro assays to evaluate the plant growth promoting abilities. Both strains were identified as Erwinia sp., demonstrating in vitro biosynthesis of IAA, ACC deaminase activity, as well as the capacity to grow in presence of NaCl and PEG. Considering the inoculation of plants, pea biometric parameters were unaffected by the presence of the bacteria, independently by the considered genotype. Conversely, the three pea genotypes differed in the regulation of antioxidant genes coding for catalase (PsCAT) and superoxide dismutase (PsSOD). The highest proline levels (212.88 μmol g−1) were detected in salt-stressed Lincoln plants inoculated with PG1, along with the up-regulation of PsSOD and PsCAT. Conversely, PG2 inoculation resulted in the lowest proline levels that were observed in Lincoln and Meraviglia d’Italia (35.39 and 23.67 μmol g−1, respectively). Overall, this study highlights the potential of these two strains as beneficial plant growth-promoting bacteria in saline environments, showing that their inoculation modulates responses in pea plants, affecting antioxidant gene expression and proline accumulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leguminous species such as pea (Pisum sativum) are an important source of nutritional components due to the seed richness in proteins, carbohydrates, vitamins, minerals, fibers and antioxidant compounds (Sapre et al. 2022). Pea can be grown in different regions, and it is fourth in legume global production after soybean, peanut, and dry beans (Vidal-Valverde et al. 2003). It is an important food and fodder legume for Mediterranean countries, including Tunisia, due to high nutritional value of their protein and starch-rich seeds (El Idrissi et al. 2020). Being a legume species, pea cultivation increases the content of nitrogenous compounds in nitrogen-poor soils (Kebede 2021). However, throughout the course of the five years from 2017 to 2021, pea output worldwide decreased by around 10% as a result of declining soil productivity and harvested area overall (FAOSTAT 2021). This scenario will be exacerbated by the increase of food demand (from 35 to 56%) globally, and the subsequent need to achieve increased crop yields under the world growing population (van Dijk et al. 2021). Addressing this challenge needs effective agricultural management to both enhance crop resilience to abiotic stresses and to preserve soil fertility and health, especially under environmental constraints (van Dijk et al. 2021).
Arid and semi-arid areas are among the most vulnerable to environmental constraints that are exacerbated by climate change. Salinity is a relevant environmental limiting element, causing major reductions in plant productivity (Peng et al. 2023; Raza et al. 2022). It is estimated that more than 50% of the arable soils in the world will be salinized by 2050 (Etesami et al. 2022). In Tunisia, about 25% of the cultivated area is affected by salinity, and it is evaluated that 9.13% of the total Tunisian surface is challenged by this threat due to aridity and poor water management (Aloui et al. 2022). The growth of crops is negatively affected by salt stress, which causes osmotic stress due to decreased water availability and ion toxicity from nutritional imbalances (Acosta-Motos et al. 2017).
Although P. sativum is recognized to be moderately tolerant to soil salinity (Shahid et al. 2022), elevated salt levels in soil were reported to significantly affect its productivity (Noreen and Ashraf 2009). In previous few years, inoculation of plants with beneficial soil microbes, including plant growth-promoting bacteria (PGPB), has proven to be a useful approach to enhance sustainable agriculture in soils subjected to several abiotic stresses, including salinity (Bhat et al. 2023, 2020; Peng et al. 2023; Mishra et al. 2021). They can increment the germination of seeds as well as the leaf size, improve chlorophyll and protein amount, increase crop growth and yield, in addition to nutrient accessibility, and delay the senescence of leaves (Saghafi et al. 2019). They ameliorate salt stress tolerance by different mechanisms, i.e., synthetizing antioxidant enzymes, non-enzymatic antioxidants, and osmolytes (e.g., proline), improving nutrient uptake, producing 1-aminocyclopropane-1-carboxylate (ACC) deaminase, indole acetic acid (IAA), siderophores and exopolysaccharide (EPS; Peng et al. 2023; Saghafi et al. 2019). Endophytic bacteria, such as Bacillus subtilis and Pseudomonas fluorescens, significantly decreased Na+ accumulation and promoted K+ uptake in pea under salt stress, affecting osmoregulation and antioxidant capacity (Sofy et al. 2021). Plant salt tolerance can be affected by the capacity of microbes to regulate the expression of plant transcription factors involved in stress responses, additionally to the synthesis of enzymes correlated to reactive oxygen species (ROS), proline and the synthesis of EPS (Aeron et al. 2020). It was observed that PGPB may improve plant development in a way that depends on the bacterial strain and plant genotype, as different plant genotypes may differentially react to the same microbial strain (Wei and Jousset 2017). Several reports have shown the potential of PGPB in improving productivity of many legume plants, including peanuts (Sharma et al. 2016), Sulla carnosa (Hidri et al. 2016) and Lathyrus cicera (Gritli et al. 2022) under salt stress conditions. Inoculation with Planomicrobium sp. MSSA-10 significantly increased pea growth under salt stress, decreasing ROS and enhancing antioxidant enzyme activities (Shahid et al. 2018). Variovorax paradoxus 5C-2 has been found to mitigate salt stress in pea, enhancing water relations, ion homeostasis and photosynthesis (Wang et al. 2016), while the application of P. sativum with Bacillus marisflavi (CHR JH 203) and Bacillus cereus (BST YS1_42) affected the expression of genes involved in ROS scavenging, defense and cell rescue, and enhanced growth and tolerance to the stress itself in greenhouse under salt stress (Gupta et al. 2021a).
Several bacterial species have been characterized as PGPB, including species belonging to genera known to be plant pathogens (Wei et al. 2023; Passera et al 2019). Among them, Pseudomonas syringae is a bacterial genus that include strains with beneficial effect on plants (i.e., P. syringae pv. syringae strain 260–02) and others that, on the contrary, are pathogenic ones (i.e., P. syringae pv. tomato strain DC3000) (Passera et al. 2019). It is known that Erwinia species may cause fire blight to Rosaceae plants (e.g., E. amylovora), and bacterial blight to cucurbits and other dicotyledonous plants, soft rot in a broad-host-range (E. carotovora), but also show positive associations with plants such as tea, rice and wheat (e.g., E. tasmaniensis and E. billingiae) (Jia et al. 2022; Sagar et al. 2018). The application of Erwinia sp. as PGPB significantly improved tomato fruit fresh and dry weight and yield compared with Bacillus pumilus and P. putida (Shen et al. 2012).
This study was aimed to verify the impact of two strains of Erwinia sp., previously isolated from pea root nodules, on alleviating salt stress in three P. sativum genotypes, using both two cultivars largely employed in Tunisia (Merveille de Kelvedon and Lincoln) and one dwarf genotype commercialized in Italy (Meraviglia d’Italia). The effects of the bacterial inoculation on pea response to a salt stress condition were evaluated by considering biometric parameters, biochemical traits, and expression of genes previously correlated with pea salt stress response.
Materials and Methods
Isolation of the bacterial strains
Soil of two fields in Tunisia served as the source of the pea nodule associated bacterial strains. Briefly pea plants (Merveille de Kelvedon cultivar) were grown using soil samples from two northern Tunisian locations (Mograne and Zaghwan) for capturing bacteria found in the native soil (Ilahi et al. 2021). For each region, three soil samples were collected at a distance of 50 m apart from each other and they were mixed to obtain a pooled sample by location. Pea seeds were sterilized and germinated as described by Gritli et al. (2019). The seedlings were grown in a greenhouse for fifty days upon the natural light with a daily temperature of 20–24 °C (minimum–maximum). At the end of the growing period, root nodules were collected and sterilized on the surface according to Vincent's standard procedures (Mnasri et al. 2009). Bacterial strains were isolated from collected root nodules by selecting individual colonies on mannitol yeast agar (YEMA) (Vincent 1970). The isolation of 20 isolates, grown on YEMA-RC (Red Congo) medium at 28 °C, allowed to distinguish two different groups of bacterial strains. The first majority group (16/20) was composed of bacteria with slow growth (appearance of colonies after four days). The second minority group (4/20) included fast-growing bacteria (appearance of colonies after two days). Two strains (called PG1 and PG2), belonging to the second group (i.e., both showing a fast growth), were selected for biochemical and molecular characterizations.
Estimation of PGP activities in vitro
Different biochemical capabilities have been assessed in PG1 and PG2 strains, regarding the inorganic phosphate solubilization, the capacity to produce indolacetic acid (IAA) and ammonia (NH3), the utilization of 1-aminocyclopropane-1-carboxylic acid (ACC), and the possibility to grow in diverse stressed conditions such as osmotic and saline environment.
To test the capacity of bacteria to solubilize inorganic phosphate, bacterial cell suspension (5 μl, 1 × 108 CFU mL−1) was spotted on plates with Pikovskaya agar medium with tri-calcium phosphate (Ca3(PO4)2), considering a triplicate for each plate. Following an incubation at 28 °C for 72 h, bacterial colonies with the clarification haloes was considere as positive for phosphate solubilization activity, and diameter of haloes were evaluated by Fiji software (ImageJ1.50i; Schneider et al. 2012).
Production of IAA was measured by using a colorimetric detection assay in liquid culture (Karnwal 2009). Isolates were cultured in Luria–Bertani (LB) broth, and then 300 μL of bacteria suspension (adjusted to 1 × 108 CFU mL−1) were inoculated in 15 mL tubes filled with 3 mL of DF (Dworkin and Foster 1958) salt minimal broth with 150 μg mL−1 of L-Tryptophan. Tubes were maintained at 28 ± 2 °C on an orbital shaker (200 rpm, 48 h) in dark conditions. One mL of cell suspensions was centrifuged (4,000 rpm, 20 min, 4 °C) and 250 μL of supernatant were combined with 1 mL of Salkowski’s reagent (1.2% FeCl3 in 37% sulphuric acid). Samples were incubated twenty minutes at room temperature, using 96-well polystyrene dishes. A FluoStar Omega microplate reader was used to measure the absorbance of bacterial suspensions (Brescia et al. 2023). IAA concentrations were determined by the preparation of a standard curve using a range of 0.5–100 μg mL−1 of pure IAA, expressing the IAA amount in µg produced by bacterial suspensions at 0.5 McFarland scale density (1.5 × 108 cells mL−1).
Evaluation of the utilization of ACC by bacterial strains (Li et al. 2011) was perforemd using 2 mL aliquots of LB cultures that were centrifuged at 8000 g for 5 min. The resulting pellets were then washed two times with 1 mL of DF medium (Dworkin and Foster 1958) and resuspended in 2 mL of DF medium with 3 mM ACC. Cultures were incubated at 28 °C, using an orbital shaker (200 rpm), for 24 h and 1 mL of each culture was then centrifuged at 8000 g for 5 min. One hundred µL of the supernatants (1:10 in DF medium) and 200 µL of each diluted supernatant, in addition to 200 µL of DF + ACC medium that was used as reference, were mixed to 400 µL of ninhydrin reagent and boiled for 30 min. Three aliquots of 180 µL for each strain as well as the reference samples were placed in the 96-well polystyrene plates, and the resulting Ruhemann’s Purple colour was evaluated by using a FluoStar Omega microplate reader (570 nm absorbance). A standard curve (in the range 0.005 and 0.05 mM) was used to assess ACC amount. In detail, the ACC-utilizing strains were grouped calculating the % of residual ACC compared with that of the DF-ACC medium without inoculation (Brescia et al. 2023). Production of ammonia (NH3) by the considered bacterial cultures were evaluated by using Nessler’s reagent (Cappuccino and Sherman 1999), following both the Abdelwahed et al. (2022) modified protocol and the original method. The two considered bacterial strains were grown in triplicate in peptone broth (10 mL) for 48 h in an incubator shaker at 30 ± 0.1 °C. After centrifugation (5 min, 6000 g), aliquots of 100 µL of each bacterial supernatant were mixed with 200 µL of Nessler’s reagent in 1.5 mL tubes and the mixture was incubated for 10 min, during which its colour turned to yellow to dark brown. After mixing, 33 µL aliquots of each reaction mixture were transferred to the wells in a 96-well plate and diluted with 198 µL of ultra pure water, and a FluoStar Omega microplate reader was utilized (450 nm absorbance) for estimating the extracellular production of NH3. The absorbance of a standard reference curve, prepared using ammonium sulphate ((NH 4)2SO4) solutions (from 50 to 400 µM), was read along with samples. In the original method, greater amounts of bacterial supernatants (1 mL), Nessler’s reagent (9 mL) were utilized, keeping the same reaction timing and dilution ratio before transferring the samples to 1-mL cuvettes for absorbance measurements.
The two strains’ capacity to grow in osmotic and saline environments were also checked. The osmotic stress conditions were produced by using polyethylene glycol (PEG 6000) (Bandeppa et al. 2015). Bacterial strains were inoculated in liquid nutrient broth with glucose (NBG) medium containing 10%, 20%, 30%, 40% and 50% PEG 6000 (100 µl of a suspension at OD600 = 1) and put in a shaking incubator set at 28 °C and 120 rpm. NBG medium without PEG 6000 was used as a control.
To evaluate the growth in salt stress conditions, colonies were streaked on nutrient glucose agar (NGA) plates containing 2.5%, 5%, 7.5% or 10% NaCl (Nautiyal et al. 2000). NGA plates without any addition of NaCl (i.e., 0.5% NaCl) were used as control. Samples were kept at 28 °C (in the dark), and bacterial growth was checked after 72 h. Number of replicates was three (n = 3) for each test. Putative pathogenicity of the two bacterial strain was also tested on tomato plants (Supplementary Information file).
Biocontrol activity of bacterial strains against phytopathogens
Selected strains were checked for their capacity to inhibit the growth of Fusarium oxysporum f. sp. lentis and Rhizoctonia solani (provided by CNR-ISPA ITEM collection, Bari, Italy, and Dr. Alessandro Infantino, CREA-DC, Roma, Italy, respectively). Six-mm-diameter mycelial plugs obtained from 6-day-old cultures of the two fungal phytopathogens were put in the middle of a 9-cm-diameter Petri dish containing potato dextrose agar (PDA). After growing the bacterial strains in 2 mL of nutrient broth (NB) on an orbital shaker (200 rpm, 24 h, 28 °C), they (once that they have reached an optical density to OD600 = 1) were streaked (10µL) at the opposite sides (about 7 cm apart) of PDA plates containing the fungus. Plates containing the sole fungal isolate were used as controls. Each bacterial strain and each fungal control were cultured in triplicate for 6 days at 25 °C. The diameters of fungal colonies were evaluated to calculate the inhibition rate (% of 1 – dt/dc, where dt is the mean test fungal colony diameter and dc is the control one).
Molecular characterization of bacterial strains
The DNA of the two considered strains (PG1 and PG2), grown overnight in NBG on a shaking incubator (28 °C, 180 rpm), was extracted using the E.Z.N.A.® Bacterial DNA Kit. Primers 27F (GAGAGTTTGATCCTGGCTCAG) (Melničáková et al. 2013) and 1495R (CTACGGCTACCTTGTTACGA) (Ventura et al. 2002) were used to amplify the 16S rRNA gene, using the protocol described in Supplementary Information file. The considered strains were also characterized for nifH encoding a nitrogenase as described in Supplementary Information file.
Seed sterilization and plant growth conditions
Seeds of two different pea genotypes commonly cultivated by Tunisian farmers were used: cv. Merveille de Kelvedon a medium-early, sweet and highly sought-after variety easy to grow, and cv. Lincoln, which is a medium-early variety, excellent for home gardens and organic production and it is adapted to the edapho-climatic conditions of Tunisia. Seeds were sterilized in 70% v/v EtOH for 60 s and in 2.5% v/v sodium hypochlorite (30 min), washed five times with sterilized water, put in Petri dishes on moist sterilized filter paper, and stored for 7 days at 25 °C (in the dark). Thirty Lincoln seedlings were transplanted to pots (0.7 L) containing sterilized quartz sand, while 30 Kelvedon and 60 Meraviglia d’Italia seedlings were transplanted to pots (1.4 L) containing sterilized quartz sand and coconut fiber (1:1 v/v). Plants were watered to compensate for evapotranspiration for one month, then twice per week with water and once a week with 50 mL ½ strength Hoagland solution (Hoagland and Arnon 1950), and they were grown in a greenhouse following the natural sun photoperiod (September–November 2022).
Bacterial inoculation in pea plants
After 28, 13 and 10 days after transplanting for Merveille de Kelvedon, Lincoln and Meraviglia d’Italia, respectively, plants were inoculated with the strains PG1 and PG2. Inoculum was prepared growing PG1 and PG2 for two days in NBG using a shaking incubator set at 26 °C (180 rpm). When PG1 and PG2 cultures reached the OD600 of 3.5, equivalent to approximately 1.9 × 109 and 1.2 × 109 CFU mL−1, plants were inoculated by pipetting 1 mL of culture in each pot at 1–2 cm depth, with pipette tips maintained at 45° to reach the root apparatus. Ten independent plants (respresenting the biological replicates) were prepared for each bacterial strain, in addition to other ten uninoculated control plants for genotypes Merveille de Kelvedon and Lincoln. Conversely, twenty independent biological replicates were used for each bacterial strain for Meraviglia d’Italia, in addition to twenty uninoculated control plants.
Salt application
After 32, 32, 23 days since transplanting, for Merveille de Kelvedon, Lincoln and Meraviglia d’Italia, respectively, 50 mL of NaCl (50 mM) starting solution was dispensed in each pot (n = 5 for Merveille de Kelvedon and Lincoln, n = 10 for Meraviglia d’Italia). Plants were subjected to salt stress watering them two times a week starting with a solution containing 100 mM NaCl and enhancing this concentration of 50 mM every three days to avoid salt shock, until reaching 200 mM NaCl as a stress condition (Pollastri et al. 2018). After 15 days from transplanting (i.e., 47, 47 and 38 days after transplanting for Merveille de Kelvedon, Lincoln and Meraviglia d’Italia, respectively) the experiment was stopped.
Biometrical and physiological measurements of plants
Before the beginning of the stress treatments, as well as during the stress period, biometric and physiological measurements were carried out. Height, node number, chlorophyll content by SPAD (Opti-Sciences) and stomatal conductance (gs) with LI-COR 600 (Ecosearch) were measured. At harvest (i.e., at the end of the stress treatment, when plants showed clear stress symptoms such as chlorosis), shoots were collected in liquid nitrogen and kept at − 80 °C for gene expression analysis and metabolic stress markers, while roots were oven-dried dried at 70 °C for 48 h for biomass determination. For Meraviglia d’Italia plants, in addition to those analyses, shoot biomass was also determined using the additional set of prepared replicates (n = 5). Pod number was recorded for all genotypes.
Biochemical analysis of P. sativum stress markers
Lipid peroxidation was evaluated through the malondialdehyde (MDA) content, resulting from the thiobarbituric acid (TBA) reaction (Heath and Packer 1968). Hydrogen peroxide (H2O2) amount was determined using the KI method (Loreto and Velikova 2001), while free proline levels were quantified using the ninhydrin reaction (Bates et al. 1973). Details have been reported in the Supplementary Information file.
Quantitative gene expression analysis in P. sativum leaves
Three biological replicates for each stress treatment (non-stressed, NS; salt stress, S) and for each inoculum (PG1, PG2, not inoculated control) were considered for the evaluation of gene expression. Total RNA was extracted from leaves (100 mg) of each biological replicate considered for gene expression (n = 3) utilizing the Spectrum Plant Total RNA extraction kit (Sigma-Aldrich) with slight modifications. RNA was quantified by NanoDrop 1000 spectrophotometer and then was treated with TURBO™ DNase kit following the routine DNase treatment (Thermo Fisher Scientific). DNA contamination absence was verified prior to proceed with the cDNA synthesis, by using pea PsPP2A specific primers in one-step RT-PCR (Knopkiewicz and Wojtaszek 2018) (Table 1). Total RNA was used to generate the cDNA, according to the SuperScript II Reverse Transcriptase® (Invitrogen) protocol using random primers. Each RT-qPCR reaction was conducted on a total volume of 10 μL, containing 1 μL diluted cDNA (dilution 1:2), 5 μL Power SYBR Green PCR Master Mix; Biorad), 3.6 μL of water and 0.2 μL of each primer (10 μM), using a 96-well plate. Primers are reported in Table 1. Reactions were performed using the Connect™ Real-Time PCR Detection System (Bio-Rad Laboratories). Thermal cycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Pea target transcripts in leaves were quantified after normalization with two reference genes: PsPP2A encoding a protein phosphatase 2A (Knopkiewicz and Wojtaszek 2018) and PsEF1α coding for an elongation factor gene (NM_001427569.1). Three independent biological replicates were used (with the exception of control of salt stressed plants of genotype Meraviglia d’Italia for which two biological replicates were considered) as well as two technical replicates for each biological one. The candidate gene expression was calculated using the equation 2−ΔΔCT (Livak et al. 2001).
Statistical analysis
R software (version 4.1.1) was used for statistical analysis. Effects of pea genotype, applied stress, and inocula, on biochemical, biometric and physiological data at the end of the experiment were tested with analyses of variance (three-way ANOVA). Differences among treatments within the plant genotypes were statistically assessed using one-way ANOVA and a Tukey’s HSD test, transforming data when necessary to fulfill ANOVA assumptions. A probability level of p-value < 0.05 was considered for all tests. Principal component analysis (PCA) was utilized for comparing both the biometric and physiological data, as well as the biochemical ones. R software (version 4.1.1) was utilized to calculate the Pearson correlation matrix. The Relative Expression Software Tool REST© 2009 v. 2.0.13 (Qiagen) (Pfaffl et al. 2002) was used for performing statistical analysis on gene expression data, using 0.05 as significance of p-value. To conduct the correlation analysis (Pearson correlation) between biochemical stress marker and gene expression (relative expression, ΔCT) data, a square root transformation was carried out to ensure normality and improve the linearity assumption. This analysis was conducted using the R package "PerformanceAnalytics". Specifically, the "chart.Correlation" function of this package was used. To assess the significance of the observed correlations, p-values were calculated for each correlation coefficient, setting it at 0.05. The p-values were obtained using the built-in functionality of the "chart.Correlation" function.
Results
Characterization of bacterial strains
Biocontrol activity showed that six-days-old co-cultures of the phytopathogenic fungus F. oxysporum f. sp. lentis with isolates PG1 and PG2 showed on average 26.4 ± 0.7% reduction in fungal growth compared with controls (represented by the sole fungal colonies), with no significant differences among bacterial isolates. On the other hand, no growth reduction was recorded when R. solani was grown in the presence of PG1 or PG2. Molecular and biochemical characterization of the two selected strains was carried out by the 16S rRNA gene sequencing and evaluation of PGP traits. The sequencing of the PCR amplicons resulted in two sequences of 1368 bp (PG1) and 1388 bp (PG2) in length. These sequences were submitted in the GenBank database (accession number OQ919144 and accession number OQ919145 for PG1 and PG2, respectively) and compared against the NCBI database (16 March 2023). The two strains showed the highest identity with Erwinia tasmaniensis, followed by other species of Erwinia (Table 2). The phylogenetic tree confirmed the BLAST outputs, clustering the two strains close to E. tasmaniensis (Fig. 1). Based on PCR performed with primers for nifH, PG2 produced an amplicon between 300 and 400 bp, while PG1 did not. The amplicon was sequenced, showing 98.78% of identity with Rhizobium sp. KNb1 nifH (e-value of 2e−119) and submitted to NCBI GenBank (accession number OR083229). Tests in pure culture showed that both bacterial strains had the capacity to produce IAA and ACC deaminase activity and that PG1 showed the capacity to solubilize phosphate (Table 3). The ability of producing extracellular NH3 by both bacterial strains could be visually assessed by observing the development of the orange colour by surnatant-based reactions mixtures after the addition of Nessler’s reagent. An apparent darker colour, suggesting higher NH3 production, was detectable in the reaction mixtures obtained from the strain PG2. Unfortunately, the absorbance data recorded from both the original method and from the 96-well plates assays of supernatants from the two bacterial cultures were very variable among replicates, even when higher dilution levels (1:8 and 1:10, in place of 1:5) were used, and showed no significant differences among strains, whose extracellular NH3 production ranged between 200 and 270 µM (not shown). In the salt stress test, PG1 was able to grow until a NaCl concentration of 7.5%, while PG2 could tolerate 10% NaCl, despite showing a reduced growth. Additionally, PG1 was able to grow on 40% PEG plates, while PG2 tolerated up to 30% PEG (Table 3).
P. sativum biometric and physiological parameter evaluation
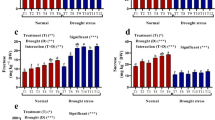
Biometric and physiological measures were taken during plant growth period, namely shoot height, node number and chlorophyll content, together with stomatal conductance, gs. Then, after harvest the pod number and pod, root and, only for Meraviglia d’Italia, shoot dry weights were checked. The analysis of variance, considering genotype, inocula and stress, showed significant differences depending on the genotype for all the considered biometric and physiological parameters, and on the applied stress for height and physiological parameters (Table S1). The PCA using biometric and eco-physiological data, recorded at the end of the experiment, suggests that three genotypes performed in a variable manner (Figure S1). Considering Merveille de Kelvedon and Lincoln, genotypes employed in Tunisia, the PCA showed a differential response to salt (Fig. 2). A significant positive correlation was found between shoot height and node number, pod dry weight and root dry weight, shoot height and root dry weight, shoot height and pod dry weight, and node number and pod dry weight (Table S2). When data from each genotype were analyzed independently, no significant differences were detected among the treatments for all the considered biometric parameters (Table S3). Concerning the physiological parameters, in Merveille de Kelvedon the chlorophyll content was significantly lower in salt stressed uninoculated and PG1-inoculated plants, compared with their non-stressed controls, while in the other two genotypes there were no statistical differences (Fig. 3). Pea plants belonging to Merveille de Kelvedon, grown without salt stress and treated with the strain PG2, had the tendency to show higher chlorophyll contents than those inoculated with PG1 (Fig. 3).
Multivariate statistical analysis of physiological (chlorophyll content and gs) and biometric (shoot height, node number, root and pod dry weight) data on Merveille de Kelvedon and Lincoln (n = 5). In (a), Principal component analysis (PCA) of samples; in (b), projection of variables, where angles are interpreted as correlations. The angle between two variable vectors represents the degree of correlation between them: adjacent (angle less than 90°) showed highly correlated variables, angle more than 90° showed uncorrelated ones
Physiological data (chlorophyll content °SPAD and gs mol m−2 s−1) in Merveille de Kelvedon (n = 5), Lincoln (n = 5) and Meraviglia d’Italia (n = 10). All data are expressed as mean ± SE (standard error). Letters are plotted on the base of Tukey’s test; different ones mean significant differences (p-value < 0.05). The absence of letters means absence of significant differences
Relating to gs values, in Merveille de Kelvedon and Lincoln the salt stressed plants showed significantly lower values than their controls, and a severe reduction of this variable under stress, irrespective of bacterial inoculation. Conversely, stomatal conductance (gs) data obtained from Meraviglia d’Italia genotype were not significantly diverse between not stressed and stressed uninoculated and PG2-inoculated plants, while PG1 bacterial inoculation led a significant difference between not stressed and stressed plants (Fig. 3).
Biochemical analysis of P. sativum stress markers
The three-way ANOVA showed significant differences for each considered factor (genotype, applied stress, inoculation) (Table S4). The PCA carried out using biochemical parameters highlighted the differential responses of the three pea genotypes: while in Merveille de Kelvedon and Lincoln plants there was a separation between not stressed and stressed plants, in Meraviglia d’Italia this separation was less evident (Fig. 4). Considering each genotype separately, salt stress affected MDA and proline quantity in Merveille de Kelvedon, significantly increasing their values, while in Lincoln and Meraviglia d’Italia only slight differences were detected between stressed and not stressed plants (Table 4). Considering bacterial inoculation, salt stressed Merveille de Kelvedon plants inoculated with PG1 consistently showed the highest values of MDA, H2O2 and proline, while in the presence of PG2 only H2O2 content was significantly enhanced compared with both uninoculated stressed plants and not stressed ones (Table 4). Lincoln plants showed a largely different behavior. Significantly higher values of MDA were detected in not stressed PG1-inoculated plants and in stressed plants treated with PG2, compared with stressed uninoculated plants that showed the lowest value. H2O2 content was similarly low in stressed plants treated with PG2 and in not stressed uninoculated plants, while the highest value was found in stressed plants inoculated with PG1. Relating to proline, the highest value was found in stressed plants inoculated with PG1 and the lowest one in not stressed plants inoculated with PG2 (Table 4). In Meraviglia d’Italia, the lowest MDA value was recorded in stressed plants treated with PG2. Conversely, significantly higher values were detected in the other stressed plants, irrespective of inoculation, and in not stressed controls. No significant differences were found among treatments for H2O2 content, while the highest proline values were detected in stressed plants inoculated with PG1, which were significantly different from those found for all the other treatments (Table 4).
Multivariate statistical analysis of biochemical data (MDA, H2O2, proline content) on the three genotypes (Merveille de Kelvedon, Lincoln and Meraviglia d’Italia). In (a), Principal component analysis (PCA) of samples; in (b), projection of variables, where angles are interpreted as correlations. The angle between two variable vectors represents the degree of correlation between them: adjacent (angle less than 90°) showed highly correlated variables, angle more than 90° showed uncorrelated ones
RT-qPCR results
To verify the plant response in the presence of salt, the expression of four genes putatively regulated by salt stress (Gupta et al. 2021a) was evaluated. The four genes include a gene coding for a superoxide dismutase (PsSOD), a catalase (PsCAT), an ascorbate peroxidase (PsAPX1) and a chlorophyll a/b binding protein (PsCHL A/B). Relative expression of PsAPX1 did not show differences among salt and/or bacterial treatments and control plants in all the genotypes, with the exception of Merveille de Kelvedon, where it was significantly up-regulated in stressed PG2-inoculated plants (Fig. 5). The transcription of PsCAT did not show differences among treatments in plants belonging to Merveille de Kelvedon genotype, while it was down-regulated in PG1-treated not stressed plants of Lincoln genotype, and up-regulated in stressed plants inoculated using the same isolate. In Meraviglia d’Italia genotype, this gene was up-regulated in salt stressed controls (p-value 0.007), stressed PG1-treated plants and in PG2-inoculated plants, while it was down-regulated in PG1-inoculated plants (Fig. 5).
Relative expression levels (Fold change ± SE), calculated following the equation 2−ΔΔCT of the four considered genes in Merveille de Kelvedon, Lincoln and Meraviglia d’Italia (n = 3, with the exception of stressed control in Meraviglia d’Italia where n = 2). Asterisks mean significant differences in comparison to control (non-stressed and uninoculated) plants, according to Pair Wise Fixed Reallocation Randomization Test
The gene PsCHL A/B was down-regulated in all the genotypes, although differently modulated by treatments. Significant down-regulation was detected in stressed Merveille de Kelvedon plants inoculated with PG1 and PG2, in all treatments of Lincoln genotype with the exception of non-stressed PG2-inoculated plants, and in PG1 treatments of both not stressed and stressed plants in Meraviglia d’Italia (Fig. 5). In Merveille de Kelvedon genotype, the PsSOD encoding gene was up-regulated only in stressed PG2-inoculated plants, while significant up-regulation in stressed controls and in PG1-inoculated and control stressed plants was detected in plants belonging to Lincoln and Meraviglia d’Italia genotypes, respectively (Fig. 5).
Gene expression and biochemical data correlation
Data obtained by gene expression and biochemical analyses were correlated, putting in evidence that in Merveille de Kelvedon and Lincoln nine correlations were detected, while in Meraviglia d’Italia only one. In detail, the significant positive correlations in Merveille de Kelvedon were between the relative expression of PsSOD and PsCAT and of PsSOD and PsAPX1 and among the three stress markers (MDA, H2O2 and proline), while the relative expression of PsCHL A/B was significantly negatively correlated with the same stress markers. The significant positive correlations in Lincoln were between the relative expression of PsSOD and PsCAT and of PsCAT and PsAPX1, these three genes with proline, PsSOD with H2O2, H2O2 with proline, while the negative correlations were between the relative expression of PsSOD and PsCHL A/B and of PsCHL A/B with proline (Fig. 6). The only positive correlation in Meraviglia d’Italia was between the relative expression of PsCAT and proline.
Correlation matrices of biochemical and target gene expression data in the three genotypes. Matrix of data in Merveille de Kelvedon (a), Lincoln (b) and Meraviglia d’Italia (c). Each diagonal subplot shows the distribution of data of the considered variable as a grey histogram. Scatterplots of each pair of variables with a least-squares reference line (red) are also reported. The slope of red line corresponds to the Pearson correlation coefficient. Numbers in the matrix represent the Pearson correlation coefficients (r). Font size used for correlation coefficients mirrors the degree of correlation. Red asterisks showed the p-value of the correlation ( = p < 0.1, * = p < 0.05, ** = p < 0.01, *** = p < 0.001)
Discussion
Plants can activate different mechanisms to protect themselves from stressful conditions, such as in high salinity (Raza et al. 2019). It has been widely reported that the application of beneficial bacteria can increase salt tolerance in different crops such as tomato, chickpea, French bean, canola, wheat, rice, maize, potato, and pepper (Gupta et al. 2022; Orozco-Mosqueda et al. 2020). Gupta et al. (2021b) recently demonstrated in a pot experiment that Bacillus subtilis RhStr_71, Bacillus safensis RhStr_223, and Bacillus cereus RhStr_JH5 improved pea plant growth, increasing the abundance of osmoprotectant and antioxidant compounds and reducing oxidative stress level due to salt. In addition, combination of PGPB and mycorrhizal fungi has been documented to mitigate the negative salt effects, enhancing Lathyrus cicero plant growth (Gritli et al. 2022).
Here, two bacteria isolated from Tunisian pea root nodules have been characterized molecularly and biochemically, and the effects of these strains on three pea genotypes in both optimal and salt stress conditions have been evaluated.
Characterization of bacterial strains
Sequencing of 16S rRNA gene allowed to verify that the two bacterial isolates both resulted to belong to Erwinia sp., phylogenetically close to Erwinia tasmaniensis. This species was first isolated from flowers and bark of apple and pear trees in Australia, showing that bacteria belonging to this species were non-phytopathogenic (Geider et al. 2006). The tests carried out by challenging tomato plants with the two isolates showed the absence of disease symptoms, confirming the non-pathogenicity of these bacterial strains, at least on two plant species (pea and tomato). The presence of non-pathogenic Erwinia species, among which E. tasmaniensis strains from different countries, was already reported (Sagar et al. 2018). Available genome sequences support these data, suggesting that non-pathogenic isolates lack the set of genetic factors needed for tissue invasion (Kube et al. 2010). Although some genetic traits involved in the induction of hypersensitive responses (HR) was still found in E. tasmaniensis strain Et1/99, able to start HR on tobacco leaves, no virulence on host apple and pear was detected (Kube et al. 2010; Palacio-Bielsa et al. 2012). The capacity of the two E. tasmaniensis strains to control the growth of a legume-derived isolate of F. oxysporum was detected in vitro, suggesting that screenings for antagonistic activity of nodule-associated microorganisms against microbial pathogens should be carried out, both in vitro and in vivo, to explore such potentially useful traits.
The molecular characterization of the bacterial strains was based on the search for the nifH gene (coding for nitrogenase), which is the most used marker gene for the identification nitrogen-fixing bacteria (Gaby and Buckley 2012). Here, its presence was detected in PG2, suggesting the potential capability of this strain to reduce nitrogen into ammonia.
On the basis of the phenotypic characterization, PG1 may represent a strain with an applicative potential due to its capacity for solubilysing phosphate, producing IAA and ACC deaminase, and to its ability to grow in presence of NaCl or PEG. The presence of these traits is common to other interesting PGPB such as Planomicrobium sp. MSSA-10 (Shahid et al. 2018), able to release organic acids in the rhizosphere resulting in increased P availability to plant roots, to regulate plant ethylene level and to produce root growth-promoting hormone. The observed features were also reported for another strain of Erwinia sp. (KP226572) that demonstrated to enhance wheat growth (Sagar et al. 2018). The bacteria producing ACC deaminase can help plant development, especially upon environmental stressful conditions (Ali et al. 2014). It was demonstrated that plants had higher levels of ethylene due to higher levels of ACC: in the presence of PGPB showing ACC deaminase activity, the ethylene levels decreased, leading to the production of α-ketobutyrate and ammonia (Gupta et al. 2022; Orozco-Mosqueda et al. 2020).
Effects of salt stress on biometric and physiological parameters of P. sativum plants
Although each genotype showed a different behavior, neither salt stress nor the presence of bacterial inoculation led to significant effects on growth parameters and root biomass. Previous studies showed that pea growth resulted negatively affected by salt treatments exceeding 50 mM and that variable responses could be detected among genotypes, depending on their tolerance (Shahid et al. 2011, 2012). A positive correlation between pod dry weight and root dry weight, and between pod dry weight and shoot height, was observed in Merveille de Kelvedon and Lincoln plants, suggesting that the tendency for more developed root and foliar systems can be associated with heavier pods. This result is in line with the fact that well-developed roots are generally expected to support nutrition, growth and development of the whole plant, as previously observed in other legume species (Gopalakrishnan et al. 2016).
Salt stress dramatically decreased the gs values in Merveille de Kelvedon and Lincoln pea genotypes, independently on the inoculation, in agreement with studies showing that reductions in stomatal conductance, along with photosynthetic rates, are common plant response to salt stress, being these variables affected by Na+/K+ ratio and turgor decline (Hernández et al. 2000; Irshad et al. 2021). A less severe gs variation was observed in Meraviglia d’Italia plants, suggesting that this genotype has a different stomatal behaviour with respect to the Tunisian cv. (Hochberg et al. 2018).
Stress marker evaluation
Three already known markers for stress, i.e., MDA, H2O2, and proline, were also evaluated. Large increases in MDA under salt stress were consistently detected in the Merveille de Kelvedon, particularly in the presence of the PG1 isolate; the same isolate also induced enhanced MDA levels in Meraviglia d’Italia stressed plants, whose uninoculated controls were unaffected by salt stress. Increase of MDA under salt stress was previously reported for different legume species, i.e., pea, bean and lentil (Noreen and Ashraf 2009; Taïbi et al. 2016; Yasir et al. 2021). High concentration of MDA mirrors membrane lipid peroxidation, but it can also be correlated to an acclimation process signal, able to activate regulatory correlated to plant defense and growth, and to protect against oxidative stresses (Morales and Munné-Bosch 2019). Conversely, low concentrations of MDA have been related to the less active lipid peroxidation in salt-tolerance mechanisms. MDA levels were significantly reduced by inoculation with the isolate PG2 in stressed Meraviglia d’Italia plants, in the absence of changes in H2O2 content. As MDA may play different, and largely unexplored, physiological roles, further studies are needed to reveal the impact of root-associated microorganisms on its accumulation. An increase of H2O2 concentration was detected in plants belonging to genotypes Merveille de Kelvedon and Lincoln under salt stress, according to studies already present in literature (Hernández et al. 2000; Irshad et al. 2021). Inoculation of salt stressed pea plants of both genotypes with PG1 increased H2O2 values, compared with uninoculated controls, while strain PG2 showed opposite effects on H2O2 levels, which were increased or decreased in salt stressed Merveille de Kelvedon or Lincoln pea plants, respectively. Thus, in Lincoln genotype the presence of PG2 bacterial strain might have reduced the plant stress status. In this regard, Neshat et al. (2022) suggested that the application of PGPR markedly decreased H2O2 levels, particularly under 100 mM salinity stress in canola plants. This reduction may be linked to the inhibition of Na+ uptake through roots and a concurrent decrease in K+ levels. Differential responses among genotypes and the lack of salt stress-related variations in H2O2 concentrations were previously reported for other pea cultivars (Noreen and Ashraf 2009). It is worth noting that, at high level, H2O2 is known to provoke oxidative damages to biomolecules, causing cell death, but at lower concentration it may work as a signaling molecule (Černý et al. 2018). The accumulation of such molecules in response to stress factors and microbial inoculants has proven highly dependent on plant and microbial identity and on their specific ability to modulate the multiple factors involved in stress response. Additionally, the regulation of this and other ROS depends on their generation and degradation, and on their neutralization rates by plant antioxidants. Results obtained on uninoculated plants of all pea genotypes confirmed that proline increases in the presence of osmotic stress. This amino acid plays an osmoprotective function, together with chaperone and antioxidant signal regulating function, whereas its specific role in response to stress in plants is still under debate (Szabados and Savoure 2010; Spormann et al. 2023). It is worth noting that Meraviglia d’Italia plants showed higher values of proline in not stressed conditions compared to the other two genotypes, and only a slight increase in proline content under stress. This result, together with the low MDA content detected in both not stressed and stressed Meraviglia d’Italia plants, may suggest a different response to salt stress for this genotype that harbors an enhanced basal level of proline compared to Merveille de Kelvedon and Lincoln. Additionally, gs data of this genotype showed lower values in not stressed conditions compared to the other two genotypes. This reduced stomatal conductance could suggest a preventive and protective strategy against water loss by Meraviglia d’Italia plants. A recent work on the combined effect of drought and salinity on forage pea plants demonstrated that tolerant pea genotypes harbored high concentration of proline (Demirkol and Yilmaz 2023). Other studies reported that the concentration of osmolytes and antioxidant enzymes in pea play an important role in salt tolerance potential (Farooq et al. 2017; Shahid et al. 2022). It is worth noting that the inoculation with the strain PG1 in Merveille de Kelvedon and Lincoln plant genotypes induced the largest accumulation of proline in response to salt stress (17- and 14-fold, respectively, over the not stressed Merveille de Kelvedon- and Lincoln-inoculated plants). It has been documented that when salt stress mitigation measures are used, including the use of PGPB, proline levels often rise (Spormann et al. 2023). However, it is still under debate if proline accumulation results from changed redox balance and hormone metabolism or if it is a component of the tolerance-inducing process (Spormann et al. 2023). In our experiment, PG2 was less able to enhance proline accumulation under stress, as only a slighter increase in stressed plants was observed compared to inoculation with PG1, suggesting that PG2 inoculated plants may not need high proline accumulation. An explanation could be that these plants have an increase activity in proline degradation or that other osmotic solutes are involved in the response to stress (Szabados and Savouré 2010).
Gene expression and correlation analysis
To cope with the oxidative damage caused by salt, plants have developed defenses based on antioxidant enzymes such as SOD, POX, CAT (Hasanuzzaman et al. 2020). Here, expression levels of these genes were strictly dependent on the considered plant genotype-bacterial strain combination. In previous works, P. sativum plants inoculated with different isolates belonging to Bacillus or Pseudomonas showed an up-regulation of the genes coding for several antioxidant enzymes in comparison to uninoculated salt-stressed plants (Gupta et al. 2021a; Sofy et al. 2021). Here, inconsistent up-regulation of PsCAT and PsSOD was detected under salt stress/bacterial treatments in genotypes Lincoln and Meraviglia d’Italia, while PsCHL A/B was down-regulated or not significant regulated, suggesting a diverse response to salt stress tolerance among pea genotypes, as already reported (El-Esawi et al. 2018; Khan et al. 2022). Correlating the gene expression and biochemical data, the highest number of significant correlations was detected in the two genotypes employed in Tunisia. In Merveille de Kelvedon, the relative expression of PsSOD was positively correlated to those of PsCAT and PsAPX1, suggesting a co-regulation of these scavenging genes. In Lincoln, the expression of PsSOD was also found to be significantly correlated with the expression of PsCAT, highlighting the co-regulation between these two functionally different genes. Additionally, always in Lincoln, a positive correlation between PsSOD expression level and the amount of H2O2 could suggest a potential relationship between the expression of this gene and oxidative stress, considering that SODs are able to catalyze the dismutation of the superoxide (O−2) radical into O2 and H2O2 (Bowler et al. 1994). A similar response to the treatments of these genes in both genotypes could be inferred. Nevertheless, looking at PsSOD and PsCAT expression on Merveille de Kelvedon and Lincoln inoculated plants, each pea genotype differentially responded to the two tested bacterial strains, with PG1 particularly affecting Lincoln stressed plants, while PG2 mainly impacted on Merveille de Kelvedon ones. In agreement with our data, a positive correlation between H2O2 level and proline content was previously found in plants subjected to environmental stress (Lee et al. 2022). The positive correlation between H2O2 and MDA in Merveille de Kelvedon could suggest a role of the peroxide in participating to lipid peroxidation and therefore to the membrane damages (Hajlaoui et al. 2009). Additionally, the correlation between MDA and proline in Merveille de Kelvedon could suggest that in presence of a cellular damage the plant adopts adaptive defensive mechanisms as osmoprotectants, as in celery under salt stress (Gao et al. 2023).
Chlorophyll a/b-binding proteins are reported to be involved in photosynthesis and to play a crucial role in capturing light energy (Bassi et al. 1997). The negative correlation between PsCHL A/B expression and the biochemical markers (MDA, proline, H2O2) in Merveille de Kelvedon, as well as between its expression and proline in Lincoln, suggested that when oxidative stress increases, and ROS rise, the expression of this gene decreases. This down-regulation may be a response to redirect cellular resources towards antioxidant defense mechanisms to cope with oxidative stress. It has been reported that reduced light absorption and increased anti-oxidative defense were associated with the down-regulation of chlorophyll a/b binding proteins (Munné-Bosch and Penuelas 2003; Xu et al. 2012). The down-regulation of PsCHL A/B in inoculated Merveille de Kelvedon and Lincoln plants under salt stress compared to uninoculated ones may also suggest that both PG1 and PG2 play a role in inducing this type of response. In Meraviglia d’Italia the only positive correlation was detected between PsCAT transcripts and proline; in upland drought tolerant rice varieties Lum et al. (2014) have already documented that their drought tolerance seemed to be correlated to activities of antioxidant enzymes, e.g., catalase, and an increase of proline content. A similar response can be hypothesized for a salt stress condition. The reduced number of correlations found in Meraviglia d’Italia could suggest that other types of responses are activated (Almeida et al. 2013), and that it could be differently affected by the applied stress.
Conclusion
The current study reveals that two non-pathogenic strains of Erwinia sp. are different in their PGP traits. By using an integrated approach, a picture of the pea plant status in three genotypes subjected to a salt stress condition was obtained and the role of the two bacterial considered Erwinia sp. strains has been highlighted. Results showed the relevance of plant genotype in determining the response to bacterial inoculants as well as the differences in the plant mechanisms activated to cope with the stress in the different plant/strain combination. Overall, this study emphasizes the importance of understanding the molecular and biochemical processes occurring in plant–microbe interactions at genotype level, and the influence on plant responses to environmental stresses. Further analyses are needed to clarify the behaviour of the three genotypes, such as the leaf water potential, and to verify the effects of the bacterial inoculation in field conditions, subjectd by an increased environmental unpredictability due to the climate change scenario.
Data availability
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
References
Abdelwahed S, Saadouli I, Kouidhi S et al (2022) A new pioneer colorimetric micro-plate method for the estimation of ammonia production by plant growth promoting rhizobacteria (PGPR). Main Group Chem 21:55–68. https://doi.org/10.3233/MGC-210077
Acosta-Motos JR, Ortuño MF, Bernal-Vicente A, Diaz-Vivancos P, Sanchez-Blanco MJ, Hernandez JA (2017) Plant responses to salt stress: adaptive mechanisms. Agronomy 7:18. https://doi.org/10.3390/agronomy7010018
Aeron A, Khare E, Jha CK et al (2020) Revisiting the plant growth-promoting rhizobacteria: lessons from the past and objectives for the future. Arch Microbiol 202:665–676. https://doi.org/10.1007/s00203-019-01779-w
Ali SZ, Sandhya V, Venkateswar Rao L (2014) Isolation and characterization of drought-tolerant ACC deaminase and exopolysaccharide-producing fluorescent Pseudomonas sp. Ann Microbiol 64:493–502. https://doi.org/10.1007/s13213-013-0680-3
Almeida P, Katschnig D, de Boer AH (2013) HKT transporters—state of the art. Int J Mol Sci 14:20359–20385. https://doi.org/10.3390/ijms141020359
Aloui M, Mahjoub A, Cheikh NB, Ludidi N, Abdelly C, Badri M (2022) Genetic variation in responses to salt stress in Tunisian populations of Medicago ciliaris. Agronomy 12:1781. https://doi.org/10.3390/agronomy12081781
Bandeppa PS, Kandpal BK (2015) Evaluation of osmotolerant rhizobacteria for alleviation of water deficit stress in mustard. Green Farming 6:590. https://doi.org/10.1016/j.plaphy.2019.08.018
Bassi R, Sandonà D, Croce R (1997) Novel aspects of chlorophyll a/b-binding proteins. Physiol Plant 100:769–779. https://doi.org/10.1111/j.1399-3054.1997.tb00004.x
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207. https://doi.org/10.1007/BF00018060
Bhat MA, Kumar V, Bhat MA et al (2020) Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front Microbiol 11:1952
Bhat MA, Mishra AK, Jan S et al (2023) Plant growth promoting rhizobacteria in plant health: a perspective study of the underground interaction. Plants 1:629. https://doi.org/10.3390/plants12030629
Bowler C, Van Camp W, Van Montagu M, Inzé D, Asada K (1994) Superoxide dismutase in plants. Crit Rev Plant Sci 13:199–218. https://doi.org/10.1080/07352689409701914
Brescia F, Sillo F, Balestrini R, Sbrana C, Zampieri E (2023) Characterization of endophytic bacteria isolated from root nodules of lentil in intercropping with durum wheat. Curr Res Microbial Sci 5:100205
Cappuccino JGS (1999) Microbiology: a laboratory manual/James G., Cappuccino and Natalie Sherman (No. 576 C3). Benjamin-Cummings Pub Co.
Černý M, Habánová H, Berka M, Luklová M, Brzobohatý B (2018) Hydrogen peroxide: its role in plant biology and crosstalk with signalling networks. Int J Mol Sci 19:2812. https://doi.org/10.3390/ijms19092812
Demirkol G, Yılmaz N (2023) Morphologically and genetically diverse forage pea (Pisum sativum var. arvense L.) genotypes under single and combined salt and drought stresses. Plant Physiol Biochem 196:880–892. https://doi.org/10.1016/j.plaphy.2023.02.041
Dworkin M, Foster JW (1958) Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol 75:592–603. https://doi.org/10.1128/jb.75.5.592-603.1958
El Idrissi M, Lamin H, Bouhnik O et al (2020) Characterization of Pisum sativum and Vicia faba microsymbionts in Morocco and definition of symbiovar viciae in Rhizobium acidisoli. Syst Appl Microbiol 43:126084. https://doi.org/10.1016/j.syapm.2020.126084
El-Esawi MA, Al-Ghamdi AA, Ali HM, Alayafi AA, Witczak J, Ahmad M (2018) Analysis of genetic variation and enhancement of salt tolerance in french pea (Pisum sativum L.). Int J Mol Sci 19:2433. https://doi.org/10.3390/ijms19082433
Etesami H, Li Z, Maathuis S, Frans JM, Cooke J (2022) The combined use of silicon and arbuscular mycorrhizas to mitigate salinity and drought stress in rice. Environ Exp Bot 10:4955. https://doi.org/10.1016/j.envexpbot.2022.104955
FAOSTAT (2021) FAO statistical databases [WWW Document]. https://www.fao.org/faostat/en/#data/QCL. Accessed 12 June 2023
Farooq M, Gogoi N, Hussain M et al (2017) Effects, tolerance mechanisms and management of salt stress in grain legumes. Plant Physiol Biochem 118:199–217. https://doi.org/10.1016/j.plaphy.2017.06.020
Gaby JC, Buckley DH (2012) A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS ONE 7:e42149. https://doi.org/10.1371/journal.pone.0042149
Gao Y, Zhang J, Wang C et al (2023) Exogenous proline enhances systemic defense against salt stress in celery by regulating photosystem, phenolic compounds, and antioxidant system. Plants 12:928. https://doi.org/10.3390/plants12040928
Geider K, Auling G, Du Z, Jakovljevic V, Jock S, Völksch B (2006) Erwinia tasmaniensis sp. nov., a non-phytopathogenic bacterium from apple and pear trees. Int J Syst Evol Microbiol 56:2937–2943. https://doi.org/10.1099/ijs.0.64032-0
Gopalakrishnan S, Vadlamudi S, Samineni S, Sameer Kumar CV (2016) Plant growth-promotion and biofortification of chickpea and pigeonpea through inoculation of biocontrol potential bacteria, isolated from organic soils. Springerplus. https://doi.org/10.1186/s40064-016-3590-6
Gritli T, Ellouze W, Chihaoui SA, Barhoumi F, Mhamdi R (2019) Genotypic and symbiotic diversity of native rhizobia-nodulating red pea (Lathyrus cicera L.) in Tunisia. Syst Appl Microbiol 43:126049. https://doi.org/10.1016/j.syapm.2019.126049
Gritli T, Boubakri H, Essahibi A, Hsouna J, Ilahi H, Didier R, Mnasri B (2022) Salt stress mitigation in Lathyrus cicera by combining different microbial inocula. Physiol Mol Biol Plants 28:1191–1206. https://doi.org/10.1007/s12298-022-01205-4
Gupta A, Bano A, Rai S, Kumar M, Ali J, Sharma S, Pathak N (2021a) ACC deaminase producing plant growth promoting rhizobacteria enhance salinity stress tolerance in Pisum sativum. 3 Biotech 11:514. https://doi.org/10.1007/s13205-021-03047-5
Gupta A, Rai BA, Khanam A, Sharma S, Pathak N (2021b) Comparative evaluation of different salt-tolerant plant growth-promoting bacterial isolates in mitigating the induced adverse effect of salinity in Pisum sativum. Biointerface Res Appl Chem 11:13141–13154. https://doi.org/10.33263/BRIAC115.1314113154
Gupta A, Mishra R, Rai S et al (2022) Mechanistic insights of plant growth promoting bacteria mediated drought and salt stress tolerance in plants for sustainable agriculture. Int J Mol Sci 23:3741. https://doi.org/10.3390/ijms23073741
Hajlaoui H, Denden M, El Ayeb N (2009) Differential responses of two maize (Zea mays L.) varieties to salt stress: changes on polyphenols composition of foliage and oxidative damages. Ind Crops Prod 30:144–151. https://doi.org/10.1016/j.indcrop.2009.03.003
Hasanuzzaman M, Bhuyan MHMB, Zulfiqar F et al (2020) Reactive oxygen species and antioxidant metabolism in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants 9:681. https://doi.org/10.3390/antiox9080681
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Int Physiol Biochim Biophys 125:189–198. https://doi.org/10.1016/0003-9861(68)90654-1
Hernández JA, Jiménez A, Mullineaux P, Sevilia F (2000) Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ 23:853–862. https://doi.org/10.1046/j.1365-3040.2000.00602.x
Hidri R, Barea JM, Mahmoud OM, Abdelly C, Azcón R (2016) Impact of microbial inoculation on biomass accumulation by Sulla carnosa provenances, and in regulating nutrition, physiological and antioxidant activities of this species under non-saline and saline conditions. J Plant Physiol 201:28–41. https://doi.org/10.1016/j.jplph.2016.06.013
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Circ Calif Agric Exp Stn 347:32
Hochberg U, Rockwell FE, Holbrook NM, Cochard H (2018) Iso/Anisohydry: a plant-environment interaction rather than a simple hydraulic trait. Trends Plant Sci 23:112–120. https://doi.org/10.1016/j.tplants.2017.11.002
Ilahi H, Hsouna J, Ellouze W et al (2021) Phylogenetic study of rhizobia nodulating pea (Pisum sativum) isolated from different geographic locations in Tunisia. Syst Appl Microbiol 44:126221. https://doi.org/10.1016/j.syapm.2021.126221
Irshad A, Rehman RNU, Abrar MM, Saeed Q, Sharif R, Hu T (2021) Contribution of rhizobium–legume symbiosis in salt stress tolerance in Medicago truncatula evaluated through photosynthesis, antioxidant enzymes, and compatible solutes accumulation. Sustainability 13:3369. https://doi.org/10.3390/su13063369
Jia H, Xi Z, Ma J, Li Y, Hag C, Lu M, Zhang ZZ, Deng WW (2022) Endophytic bacteria from the leaves of two types of albino tea plants, indicating the plant growth promoting properties. Plant Growth Regul 96:331–343. https://doi.org/10.1007/s10725-021-00779-5
Karnwal A (2009) Production of indole acetic acid by fluorescent Pseudomonas in the presence of L-tryptophan and rice root exudates. J Plant Pathol 19:61–63. https://doi.org/10.4454/jpp.v91i1.624
Kebede E (2021) Contribution, utilization, and improvement of legumes-driven biological nitrogen fixation in agricultural systems. Front Sust Food Syst 5:767998. https://doi.org/10.3389/fsufs.2021.767998
Khan MAH, Baset Mia MA, Quddus MA et al (2022) Salinity-induced physiological changes in pea (Pisum sativum L.): germination rate, biomass accumulation, relative water content, seedling vigor and salt tolerance index. Plants 11:3493. https://doi.org/10.3390/plants11243493
Knopkiewicz M, Wojtaszek P (2018) Validation of reference genes for gene expression analysis using quantitative polymerase chain reaction in pea lines (Pisum sativum) with different lodging susceptibility. Ann Appl Biol 174:86–91. https://doi.org/10.1111/aab.12475
Kube M, Migdoll AM, Gehring I et al (2010) Genome comparison of the epiphytic bacteria Erwinia billingiae and E. tasmaniensis with the pear pathogen E. pyrifoliae. BMC Genom 11:393. https://doi.org/10.1186/1471-2164-11-393
Lee BR, La VH, Park SH, Mamun MA, Bae DW, Kim TH (2022) H2O2-responsive hormonal status involves oxidative burst signaling and proline metabolism in rapeseed leaves. Antioxidants 11:566. https://doi.org/10.3390/antiox11030566
Li Z, Chang S, Lin L, Li Y, An Q (2011) A colorimetric assay of 1-aminocyclopropane-1-carboxylate (ACC) based on ninhydrin reaction for rapid screening of bacteria containing ACC deaminase. Lett Appl Microbiol 53:178–185. https://doi.org/10.1111/j.1472-765X.2011.03088.x
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Loreto F, Velikova V (2001) Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol 127:1781–1787. https://doi.org/10.1104/pp.010497
Lum MS, Hanafi MM, Rafii YM, Akmar ASN (2014) Effect of drought stress on growth, proline and antioxidant enzyme activities of upland rice. J Anim Plant Sci 24:1487–1493
Melničáková J, Derdáková M, Barák I (2013) A system to simultaneously detect tick-borne pathogens based on the variability of the 16s ribosomal genes. Parasit Vectors 6:269. https://doi.org/10.1186/1756-3305-6-269
Mishra P, Mishra J, Arora NK (2021) Plant growth promoting bacteria for combating salinity stress in plants–Recent developments and prospects: a review. Microbiol Res 252:126861
Mnasri B, Badri Y, Saïdi S, de Lajudie P, Mhamdi R (2009) Symbiotic diversity of Ensifer meliloti strains recovered from various legume species in Tunisia. Syst Appl Microbiol 32:583–592. https://doi.org/10.1016/j.syapm.2009.07.007
Morales M, Munné-Bosch S (2019) Malondialdehyde: facts and artifacts. Plant Physiol 180:1246–1250. https://doi.org/10.1104/pp.19.00405
Munné-Bosch S, Penuelas J (2003) Photo-and antioxidative protection, and a role for salicylic acid during drought and recovery in field-grown Phillyrea angustifolia plants. Planta 217:758–766. https://doi.org/10.1007/s00425-003-1037-0
Nautiyal CS, Bhadauria S, Kumar P, Lal H, Mondal R, Verma D (2000) Stress induced phosphate solubilization in bacteria isolated from alkaline soils. FEMS Microbiol Lett 182:291–296. https://doi.org/10.1111/j.1574-6968.2000.tb08910.x
Neshat M, Abbasi A, Hosseinzadeh A, Sarikhani MR, Dadashi Chavan D, Rasoulnia A (2022) Plant growth promoting bacteria (PGPR) induce antioxidant tolerance against salinity stress through biochemical and physiological mechanisms. Physiol Mol Biol Plants 28:347–361. https://doi.org/10.1007/s12298-022-01128-0
Noreen Z, Ashraf M (2009) Assessment of variation in antioxidative defense system in salt-treated pea (Pisum sativum) cultivars and its putative use as salinity tolerance markers. J Plant Physiol 166:1764–1774. https://doi.org/10.1016/j.jplph.2009.05.005
Orozco-Mosqueda MDC, Glick BR, Santoyo G (2020) ACC deaminase in plant growth-promoting bacteria (PGPB): an efficient mechanism to counter salt stress in crops. Microbiol Res 235:126439. https://doi.org/10.1016/j.micres.2020.126439
Palacio-Bielsa A, Roselló M, Llop P, López MM (2012) Erwinia spp. from pome fruit trees: similarities and differences among pathogenic and non-pathogenic species. Trees 26:13–29. https://doi.org/10.1007/s00468-011-0644-9
Passera A, Compant S, Casati P et al (2019) Not just a pathogen? Description of a plant-beneficial Pseudomonas syringae strain. Front Microbiol 10:1409. https://doi.org/10.3389/fmicb.2019.01409
Peng M, Jiang Z, Zhou F, Wang Z (2023) From salty to thriving: plant growth promoting bacteria as nature’s allies in overcoming salinity stress in plants. Front Microbiol 14:1169809. https://doi.org/10.3389/fmicb.2023.1169809
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. https://doi.org/10.1093/nar/30.9.e36
Pollastri S, Savvides A, Pesando M et al (2018) Impact of two arbuscular mycorrhizal fungi on Arundo donax L. response to salt stress. Planta 24:573–585. https://doi.org/10.1007/s00425-017-2808-3
Raza A, Razzaq A, Mehmood SS, Zou X, Zhang X, Lv Y, Xu J (2019) Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants 8:34. https://doi.org/10.3390/plants8020034
Raza A, Tabassum J, Fakhar AZ et al (2022) Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit Rev Biotechnol 15:1–28. https://doi.org/10.1080/07388551.2022.2093695
Sagar A, Thomas G, Rai S, Mishra RK, Ramteke PW (2018) Enhancement of growth and yield parameters of wheat variety AAI-W6 by an organic farm isolate of plant growth promoting Erwinia Species (KP226572). Int J Environ Agric Biotechnol 11:159–171. https://doi.org/10.30954/0974-1712.2018.00178.21
Saghafi D, Delangiz N, Lajayer BA, Ghorbanpour M (2019) An overview on improvement of crop productivity in saline soils by halotolerant and halophilic PGPRs. 3 Biotech 9:261. https://doi.org/10.1007/s13205-019-1799-0
Sapre S, Gontia-Mishra I, Tiwari S (2022) Plant growth-promoting rhizobacteria ameliorates salinity stress in pea (Pisum sativum). J Plant Growth Regul 41:647–656. https://doi.org/10.1007/s00344-021-10329-y
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. https://doi.org/10.1038/nmeth.2089
Shahid M, Pervez M, Balal R et al (2011) Effect of salt stress on growth, gas exchange attributes and chlorophyll contents of pea (Pisum sativum). Afr J Agric Res 6:5808–5816
Shahid M, Balal R, Pervez M et al (2012) Differential response of pea (Pisum sativum L.) genotypes to salt stress in relation to the growth, physiological attributes antioxidant activity and organic solutes. Aust J Crop Sci 6:828–838
Shahid M, Akram MS, Khan MA et al (2018) A phytobeneficial strain Planomicrobium sp. MSSA-10 triggered oxidative stress responsive mechanisms and regulated the growth of pea plants under induced saline environment. J Appl Microbiol 124:1566–1579. https://doi.org/10.1111/jam.13732
Shahid S, Shahbaz M, Maqsood MF et al (2022) Proline-induced modifications in morpho-physiological, biochemical and yield attributes of pea (Pisum sativum L.) cultivars under salt stress. Sustainability 14:13579. https://doi.org/10.3390/su142013579
Sharma S, Kulkarni J, Jha B (2016) Halotolerant rhizobacteria promote growth and enhance salinity tolerance in peanut. Front Microbiol 7:1600. https://doi.org/10.3389/fmicb.2016.01600
Shen M, Jun Kang Y, Li Wang H, Sheng Zhang X, Xin Zhao Q (2012) Effect of plant growth-promoting rhizobacteria (PGPRs) on plant growth, yield, and quality of tomato (Lycopersicon esculentum Mill.) under simulated seawater irrigation. J Gen Appl Microbiol 58:253–262. https://doi.org/10.2323/jgam.58.253
Sofy MR, Aboseidah AA, Heneidak SA, Ahmed HR (2021) ACC deaminase containing endophytic bacteria ameliorate salt stress in Pisum sativum through reduced oxidative damage and induction of antioxidative defense systems. Environ Sci Pollut Res Int 28:40971–40991. https://doi.org/10.1007/s11356-021-13585-3
Spormann S, Nadais P, Sousa F et al (2023) Accumulation of proline in plants under contaminated soils- are we on the same page? Antioxidants 12:666. https://doi.org/10.3390/antiox12030666
Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15:89–97. https://doi.org/10.1016/j.tplants.2009.11.009
Taïbi K, Taïbi F, Abderrahim LA, Ennajah A, Belkhodja M, Mulet JM (2016) Effect of salt stress on growth, chlorophyll content, lipid peroxidation and antioxidant defence systems in Phaseolus vulgaris L. South Afric J Bot 105:306–312. https://doi.org/10.1016/j.sajb.2016.03.011
van Dijk M, Morley T, Rau ML, Saghai Y (2021) A meta-analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nat Food 2:494–501. https://doi.org/10.1038/s43016-021-00322-9
Ventura M, Zink R (2002) Specific identification and molecular typing analysis of Lactobacillus johnsonii by using PCR-based methods and pulsed-field gel electrophoresis. FEMS Microbiol Lett 217:141–154. https://doi.org/10.1111/j.1574-6968.2002.tb11468.x
Vidal-Valverde C, Frias J, Hernandex A et al (2003) Assessment of nutritional compounds and antinutritional factors in pea (Pisum sativum) seeds. J Sci Food Agric 83:298–306. https://doi.org/10.1002/jsfa.1309
Vincent JM (1970) A manual for the practical study of the root-nodule bacteria. In: IBP handbook 15, Blackwell, Oxford and Edinburgh
Wang Q, Dodd IC, Belimov AA, Jiang F (2016) Rhizosphere bacteria containing 1-aminocyclopropane-1- carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na+ accumulation. Funct Plant Biol 43:161–172. https://doi.org/10.1071/FP15200
Wei Z, Jousset A (2017) Plant breeding goes microbial. Trends Plant Sci 22:555–558. https://doi.org/10.1016/j.tplants.2017.05.009
Wei X, Moreno-Hagelsieb G, Glick BR, Doxey AC (2023) Comparative analysis of adenylate isopentenyl transferase genes in plant growth-promoting bacteria and plant pathogenic bacteria. Heliyon 9:e13955. https://doi.org/10.1016/j.heliyon.2023.e13955
Xu YH, Liu R, Yan L et al (2012) Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J Exp Bot 63:1095–1106. https://doi.org/10.1093/jxb/err315
Yasir TA, Khan A, Skalicky M et al (2021) Exogenous sodium nitroprusside mitigates salt stress in lentil (Lens culinaris Medik.) by affecting the growth, yield, and biochemical properties. Molecules 26:2576. https://doi.org/10.3390/molecules26092576
Acknowledgements
This research was supported by the PRIMA RESCHEDULE project (Italian MUR DD 1293/2021) and the Tunisian-Moroccan bilateral project (20/PRD03). This study was carried out within the Agritech National Research Center and received funding from the European Union Next-Generation EU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR) – MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4 – D.D. 1032 17/06/2022, CN00000022). This manuscript reflects only the authors’ views and opinions, neither the European Union nor the European Commission can be considered responsible for them.
Funding
Open access funding provided by Consiglio Nazionale Delle Ricerche (CNR) within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
RB, BM: conceptualization; HI, CS, FB, MMEI, MNA, LO: isolation and biochemical bacterial characterization; HI, EZ, LG, MS, AC: greenhouse experiment; HI, EZ, FB, MS: molecular characterization and gene expression analysis; RM, GG, VF: stress markers characterization; EZ, LG, AC, FS: statistical analysis; HI, EZ: writing original draft; EZ, CS, FB, GG, MS, FS, VF, RB, BM: review and editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12298_2024_1419_MOESM1_ESM.pdf
Figure S1 Principal component analysis of biometric and physiological parameters (shoot height, node number, root and pod dry weight, chlorophyll content, and gs) performed with R (v 4.1.1) on the three genotypes (Merveille de Kelvedon, Lincoln and Meraviglia d’Italia) (n = 5 in Merveille de Kelvedon and Lincoln, while n = 10 in Meraviglia d’Italia. In (a), Principal component analysis (PCA) of samples; in (b), projection of variables, where angles are interpreted as correlations. The angle between two variable vectors represents the degree of correlation between them: adjacent (angle less than 90°) showed highly correlated variables, angle more than 90° showed uncorrelated ones. (PDF 24 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ilahi, H., Zampieri, E., Sbrana, C. et al. Impact of two Erwinia sp. on the response of diverse Pisum sativum genotypes under salt stress. Physiol Mol Biol Plants 30, 249–267 (2024). https://doi.org/10.1007/s12298-024-01419-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-024-01419-8