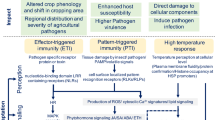
Abstract
Rice, a critical cereal crop, grapples with productivity challenges due to its inherent sensitivity to low temperatures, primarily during the seedling and booting stages. Recognizing the polygenic complexity of cold stress signaling in rice, a meta-analysis was undertaken, focusing on 20 physiological traits integral to cold tolerance. This initiative allowed the consolidation of genetic data from 242 QTLs into 58 meta-QTLs, thereby significantly constricting the genetic and physical intervals, with 84% of meta-QTLs (MQTLs) being reduced to less than 2 Mb. The list of 10,505 genes within these MQTLs, was further refined utilizing expression datasets to pinpoint 46 pivotal genes exhibiting noteworthy differential regulation during cold stress. The study underscored the presence of several TFs such as WRKY, NAC, CBF/DREB, MYB, and bHLH, known for their roles in cold stress response. Further, ortho-analysis involving maize, barley, and Arabidopsis identified OsWRKY71, among others, as a prospective candidate for enhancing cold tolerance in diverse crop plants. In conclusion, our study delineates the intricate genetic architecture underpinning cold tolerance in rice and propounds significant candidate genes, offering crucial insights for further research and breeding strategies focused on fortifying crops against cold stress, thereby bolstering global food resilience.





Similar content being viewed by others
References
Andaya VC, Mackill DJ (2003) Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. J Exp Bot 54(392):2579–2585
Agalou A, Purwantomo S, Övernäs E et al (2007) A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol 66(1–2):87–103. https://doi.org/10.1007/s11103-007-9255-7
Agarwal M, Hao Y, Kapoor A et al (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281(49):37636–37645. https://doi.org/10.1074/jbc.m605895200
Andaya VC, Tai TH (2006) Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theor Appl Genet 113(3):467–475. https://doi.org/10.1007/s00122-006-0311-5
Andaya VC, Tai TH (2007) Fine mapping of the qCTS4 locus associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Breed 20(4):349–358. https://doi.org/10.1007/s11032-007-9096-8
Arcade A, Labourdette A, Falque M et al (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20(14):2324–2326. https://doi.org/10.1093/bioinformatics/bth230
Baruah AR, Ishigo-Oka N, Adachi et al (2008) Cold tolerance at the early growth stage in wild and cultivated rice. Euphytica 165(3):459–470. https://doi.org/10.1007/s10681-008-9753-y
Bhattacharjee A, Khurana JP, Jain M (2016) Characterization of rice homeobox genes, OsHOX22 and OsHOX24, and over-expression of OsHOX24 in transgenic Arabidopsis suggest their role in abiotic stress response. Front Plant Sci. https://doi.org/10.3389/fpls.2016.00627
Boro P, Sultana A, Mandal K, Chattopadhyay S (2022) Interplay between glutathione and mitogen-activated protein kinase 3 via transcription factor WRKY40 under combined osmotic and cold stress in Arabidopsis. J Plant Physiol 271:153664. https://doi.org/10.1016/j.jplph.2022.153664
Buti M, Baldoni E, Formentin E et al (2019) A meta-analysis of comparative transcriptomic data reveals a set of key genes involved in the tolerance to abiotic stresses in rice. Int J Mol Sci 20(22):5662. https://doi.org/10.3390/ijms20225662
Chawade A, Lindlöf A, Olsson B, Olsson O (2013) Global expression profiling of low temperature induced genes in the chilling tolerant Japonica rice Jumli Marshi. PLoS ONE 8(12):e81729. https://doi.org/10.1371/journal.pone.0081729
Cruz RPD, Sperotto RA, Cargnelutti D et al (2013) Avoiding damage and achieving cold tolerance in rice plants. Food Energy Secur 2(2):96–119. https://doi.org/10.1002/fes3.25
da Cruz RP, Milach SCK (2000) Breeding for cold tolerance in irrigated rice. Ciencia Rural 30(5):909–917. https://doi.org/10.1590/S0103-84782000000500031
Dong MA, Farré EM, Thomashow MF (2011) Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci 108(17):7241–7246. https://doi.org/10.1073/pnas.1103741108
Fang C, Zhang P, Jian X et al (2017) Overexpression of Lsi1 in cold-sensitive rice mediates transcriptional regulatory networks and enhances resistance to chilling stress. Plant Sci 262:115–126. https://doi.org/10.1016/j.plantsci.2017.06.002
Filyushin MA, Kochieva EZ, Shchennikova AV (2022) ZmDREB2.9 gene in Maize (Zea mays L.): genome-wide identification, characterization, expression, and stress response. Plants 11(22):3060. https://doi.org/10.3390/plants11223060
Ge SX, Jung D, Yao R (2020) ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36(8):2628–2629. https://doi.org/10.1093/bioinformatics/btz931
Guo H, Wu T, Li S, He Q, Yang Z, Zhang W, Deng H (2019) The methylation patterns and transcriptional responses to chilling stress at the seedling stage in rice. Int J Mol Sci 20(20):5089
Hu W, Ren Q, Chen Y et al (2021) Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. https://doi.org/10.1186/s12870-021-03206-z
IRRI (2023). https://www.irri.org/our-work/impact-challenges/climate-change-sustainability
Jiang Y, Yang B, Deyholos MK (2009) Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol Genet Genomics 282(5):503–516. https://doi.org/10.1007/s00438-009-0481-3
Katiyar A, Smita S, Lenka SK et al (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics. https://doi.org/10.1186/1471-2164-13-544
Kidokoro S, Hayashi K, Haraguchi H et al (2021) Posttranslational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression in Arabidopsis. Proc Natl Acad Sci 118(10):234. https://doi.org/10.1073/pnas.2021048118
Kidokoro S, Shinozaki K, Yamaguchi-Shinozaki K (2022) Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci 27(9):922–935. https://doi.org/10.1016/j.tplants.2022.01.008
Kim CY, Vo KTX et al (2016) Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol Rep 10(1):13–23. https://doi.org/10.1007/s11816-015-0383-2
Kim SH, Kim HS, Bahk et al (2017) Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucl Acids Res 45(11):6613–6627. https://doi.org/10.1093/nar/gkx417
Kong W, Zhang C, Qiang Y et al (2020) Integrated RNA-seq analysis and meta-QTLs mapping provide insights into cold stress response in rice seedling roots. Int J Mol Sci 21(13):4615. https://doi.org/10.3390/ijms21134615
Kreps JA, Wu Y, Chang HS et al (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold Stress. Plant Physiol 130(4):2129–2141. https://doi.org/10.1104/pp.008532
Kumari A, Sharma D, Sharma P et al (2023) Meta-QTL and haplo-pheno analysis reveal superior haplotype combinations associated with low grain chalkiness under high temperature in rice. Front Plant Sci 14:234. https://doi.org/10.3389/fpls.2023.1133115
Leng Y, Sun J, Wang J et al (2020) Genome-wide lncRNAs identification and association analysis for cold-responsive genes at the booting stage in rice (Oryza sativa L.). Plant Genome. https://doi.org/10.1002/tpg2.20020
Lescot M (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Li J, Khatab AA, Hu L, Zhao L, Yang J, Wang L, Xie G (2022) Genome-wide association mapping identifies new candidate genes for cold stress and chilling acclimation at seedling stage in rice (Oryza sativa L). Int J Mol Sci 23(21):13208
Li HW, Zang BS, Deng XW et al (2011) Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234(5):1007–1018. https://doi.org/10.1007/s00425-011-1458-0
Ma Y, Dai X, Xu Y et al (2015) COLD1 confers chilling tolerance in rice. Cell 162(1):222. https://doi.org/10.1016/j.cell.2015.06.046
Marè C, Mazzucotelli E, Crosatti C et al (2004) Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol 55(3):399–416. https://doi.org/10.1007/s11103-004-0906-7
Matsukura S, Mizoi J, Yoshida T et al (2010) Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics 283(2):185–196. https://doi.org/10.1007/s00438-009-0506-y
Moraes de Freitas GP, Basu S, Ramegowda V et al (2019) Physiological and transcriptional responses to low-temperature stress in rice genotypes at the reproductive stage. Plant Signal Behav 14(4):e1581557. https://doi.org/10.1080/15592324.2019.1581557
Nah G, Lee M, Kim DS et al (2016) Transcriptome analysis of Spartina Pectinata in response to freezing stress. PLoS ONE 11(3):e0152294. https://doi.org/10.1371/journal.pone.0152294
Niggeweg R, Thurow C, Kegler C et al (2000) Tobacco transcription factor TGA2.2 Is the main component of as-1-binding factor ASF-1 and Is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J Biol Chem 275(26):19897–19905. https://doi.org/10.1074/jbc.m909267199
Pan Y, Zhang H, Zhang D et al (2015) Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS ONE 10(3):e0120590. https://doi.org/10.1371/journal.pone.0120590
Pandit E, Tasleem S, Barik SR et al (2017) Genome-wide association mapping reveals multiple QTLs governing tolerance response for seedling stage chilling stress in Indica rice. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00552
Park MR, Yun KY, Mohanty B et al (2010) Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ 33(12):2209–2230. https://doi.org/10.1111/j.1365-3040.2010.02221.x
Pasquali G, Biricolti S, Locatelli F et al (2008) Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep 27(10):1677–1686. https://doi.org/10.1007/s00299-008-0587-9
Pino MT, Skinner JS, Park EJ et al (2007) Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol J 5(5):591–604. https://doi.org/10.1111/j.1467-7652.2007.00269.x
Pradhan SK, Nayak DK, Guru M et al (2015) Screening and classification of genotypes for seedling-stage chilling stress tolerance in rice and validation of the trait using SSR markers. Plant Genet Resour 14(3):173–182. https://doi.org/10.1017/s1479262115000192
Pradhan SK, Barik SR, Sahoo A et al (2016) Population structure, genetic diversity and molecular marker-trait association analysis for high temperature stress tolerance in rice. PLoS ONE 11(8):e0160027. https://doi.org/10.1371/journal.pone.0160027
Pradhan SK, Pandit E, Nayak DK et al (2019) Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol 19(1):234. https://doi.org/10.1186/s12870-019-1922-8
Raudvere U, Kolberg L, Kuzmin I et al (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucl Acids Res 47(W1):W191–W198. https://doi.org/10.1093/nar/gkz369
Ritonga FN, Ngatia JN, Wang Y, Khoso MA et al (2021) AP2/ERF, an important cold stress-related transcription factor family in plants: a review. Physiol Mol Biol Plants 27(9):1953–1968. https://doi.org/10.1007/s12298-021-01061-8
Sakai H, Lee SS, Tanaka T et al (2013) Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol 54(2):e6–e6. https://doi.org/10.1093/pcp/pcs183
Seo JS, Joo J, Kim MJ et al (2011) OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J 65(6):907–921. https://doi.org/10.1111/j.1365-313x.2010.04477.x
Shakiba E, Edwards JD, Jodari F et al (2017) Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE 12(3):e0172133. https://doi.org/10.1371/journal.pone.0172133
Shimoyama N, Johnson M, Beaumont A et al (2020) Multiple cold tolerance trait phenotyping reveals shared quantitative trait Loci in Oryza sativa. Rice. https://doi.org/10.1186/s12284-020-00414-3
Singh K (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5(5):430–436. https://doi.org/10.1016/s1369-5266(02)00289-3
Song Y, Jiang M, Zhang H et al (2021) Zinc oxide nanoparticles alleviate chilling stress in rice (Oryza Sativa L.) by regulating antioxidative system and chilling response transcription factors. Molecules 26(8):2196. https://doi.org/10.3390/molecules26082196
Sosnowski O, Charcosset A, Joets J (2012) BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28(15):2082–2083. https://doi.org/10.1093/bioinformatics/bts313
Suzuki K, Aoki N, Matsumura H et al (2015) Cooling water before panicle initiation increases chilling-induced male sterility and disables chilling-induced expression of genes encoding OsFKBP65 and heat shock proteins in rice spikelets. Plant Cell Environ 38(7):1255–1274. https://doi.org/10.1111/pce.12498
Takesawa T, Ito M, Kanzaki H et al (2002) Over-expression of ζ glutathione S-transferase in trans-genic rice enhances germination and growth at low temperature. Mol Breed 9:93–101. https://doi.org/10.1023/A:1026718308155
Tang J, Tian X, Mei E, He M, Gao J, Yu J, Bu Q (2022) WRKY53 negatively regulates rice cold tolerance at the booting stage by fine-tuning anther gibberellin levels. The Plant Cell 34(11):4495–4515
Tao Z, Kou Y, Liu H et al (2011) OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 62(14):4863–4874. https://doi.org/10.1093/jxb/err144
Temnykh S, DeClerck G, Lukashova A et al (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11(8):1441–1452. https://doi.org/10.1101/gr.184001
Thom R, Dixon DP, Edwards R et al (2001) The structure of a zeta class glutathione S-transferase from Arabidopsis thaliana: characterisation of a GST with novel active-site architecture and a putative role in tyrosine catabolism. J Mol Biol 308(5):949–962. https://doi.org/10.1006/jmbi.2001.4638
Thurow C, Schiermeyer A, Krawczyk S et al (2005) Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J 44(1):100–113. https://doi.org/10.1111/j.1365-313x.2005.02513.x
Tian T, Liu Y, Yan H et al (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucl Acids Res 45(W1):W122–W129. https://doi.org/10.1093/nar/gkx382
Van Eck L, Davidson RM et al (2014) The transcriptional network of WRKY53 in cereals links oxidative responses to biotic and abiotic stress inputs. Funct Integr Genomics 14(2):351–362. https://doi.org/10.1007/s10142-014-0374-3
Vannini C, Locatelli F, Bracale M et al (2004) Overexpression of the riceOsmyb4gene increases chilling and freezing tolerance ofArabidopsis thaliana plants. Plant J 37(1):115–127. https://doi.org/10.1046/j.1365-313x.2003.01938.x
Veyrieras JB, Goffinet B, Charcosset A (2007) MetaQTL: a package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinform. https://doi.org/10.1186/1471-2105-8-49
Wang X, Niu QW, Teng C et al (2008) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19(2):224–235. https://doi.org/10.1038/cr.2008.276
Wang Q, Tang J, Han B et al (2019) Advances in genome-wide association studies of complex traits in rice. Theor Appl Genet 133(5):1415–1425. https://doi.org/10.1007/s00122-019-03473-3
Wang Z, Wang S, Yu C et al (2020) QTL analysis of rice photosynthesis-related traits under the cold stress across multi-environments. Euphytica. https://doi.org/10.1007/s10681-020-02651-5
Wei X, Liu S, Sun C et al (2021) Convergence and divergence: signal perception and transduction mechanisms of cold stress in Arabidopsis and rice. Plants 10(9):1864. https://doi.org/10.3390/plants10091864
Wei PL, Huang CY, Chang et al (2023) PCTAIRE protein kinase 1 (PCTK1) suppresses proliferation, stemness, and chemoresistance in colorectal cancer through the BMPR1B-Smad1/5/8 signaling pathway. Int J Mol Sci 24(12):10008. https://doi.org/10.3390/ijms241210008
Xia C, Gong Y, Chong K et al (2020) Phosphatase OsPP2C27 directly dephosphorylates OsMAPK3 and OsbHLH002 to negatively regulate cold tolerance in rice. Plant Cell Environ 44(2):491–505. https://doi.org/10.1111/pce.13938
Xiang C, Miao ZH, Lam E (1996) Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl-jasmonate and hydrogen peroxide. Plant Mol Biol 32(3):415–426. https://doi.org/10.1007/bf00019093
Xiao N, Huang WN, Li AH et al (2014a) Fine mapping of the qLOP2 and qPSR2-1 loci associated with chilling stress tolerance of wild rice seedlings. Theor Appl Genet 128(1):173–185. https://doi.org/10.1007/s00122-014-2420-x
Xiao N, Huang WN, Zhang XX et al (2014b) Fine mapping of qRC10-2, a quantitative trait locus for cold tolerance of rice roots at seedling and mature stages. PLoS ONE 9(5):e96046. https://doi.org/10.1371/journal.pone.0096046
Xie Z, Zhang ZL, Zou X et al (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137(1):176–189. https://doi.org/10.1104/pp.104.054312
Xie Z, Zhang ZL, Zou X et al (2006) Interactions of two abscisic-acid inducedWRKYgenes in repressing gibberellin signaling in aleurone cells. Plant J 46(2):231–242. https://doi.org/10.1111/j.1365-313x.2006.02694.x
Xie Y, Chen P, Yan Y et al (2017) An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol 218(1):201–218. https://doi.org/10.1111/nph.14952
Xie H, Zhu M, Yu Y et al (2022) Comparative transcriptome analysis of the cold resistance of the sterile rice line 33S. PLoS ONE 17(1):e0261822. https://doi.org/10.1371/journal.pone.0261822
Xiong J, Chen D, Chen Y et al (2023) Genome-wide association mapping and transcriptomic analysis reveal key drought-responding genes in barley seedlings. Curr Plant Biol 33:100277. https://doi.org/10.1016/j.cpb.2023.100277
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57(1):781–803. https://doi.org/10.1146/annurev.arplant.57.032905.105444
Yang Z, Huang D, Tang W et al (2013) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS ONE 8(7):e68433. https://doi.org/10.1371/journal.pone.0068433
Yang X, Wang X, Ji L et al (2015) Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep 34(6):943–958. https://doi.org/10.1007/s00299-015-1756-2
Yang T, Zhang S, Zhao J et al (2016) Identification and pyramiding of QTLs for cold tolerance at the bud bursting and the seedling stages by use of single segment substitution lines in rice (Oryza sativa L.). Mol Breed. https://doi.org/10.1007/s11032-016-0520-9
Yang LM, Liu HL, Lei L et al (2017) Identification of QTLs controlling low-temperature germinability and cold tolerance at the seedling stage in rice (Oryza Sativa L.). Euphytica. https://doi.org/10.1007/s10681-017-2092-0
Yang LM, Liu HL, Zhao HW et al (2019) Mapping quantitative trait loci and meta-analysis for cold tolerance in rice at booting stage. Euphytica. https://doi.org/10.1007/s10681-019-2410-9
Yang J, Li D, Liu H et al (2020) Identification of QTLs involved in cold tolerance during the germination and bud stages of rice (Oryza sativa L.) via a high-density genetic map. Breed Sci 70(3):292–302. https://doi.org/10.1270/jsbbs.19127
Yang L, Lei L, Li P et al (2021) Identification of candidate genes conferring cold tolerance to rice (Oryza sativa L.) at the bud-bursting stage using bulk segregant analysis sequencing and linkage mapping. Front Plant Sci. https://doi.org/10.3389/fpls.2021.647239
Yoshida R, Kanno A, Kameya T (1996) Cool temperature-induced chlorosis in rice plants (II. Effects of cool temperature on the expression of plastid-encoded genes during shoot growth in darkness). Plant Physiol 112(2):585–590. https://doi.org/10.1104/pp.112.2.585
Yu Y, Ouyang Y, Yao W (2017) shinyCircos: an R/Shiny application for interactive creation of Circos plot. Bioinformatics 34(7):1229–1231. https://doi.org/10.1093/bioinformatics/btx763
Yuan R, Zhao N, Usman B et al (2020) Development of chromosome segment substitution lines (CSSLs) derived from Guangxi wild rice (Oryza rufipogon Griff.) under rice (Oryza sativa L.) background and the identification of QTLs for plant architecture, Agronomic traits and cold tolerance. Genes 11(9):980. https://doi.org/10.3390/genes11090980
Zhan X, Wang B, Li H et al (2012) Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci 109(44):18198–18203. https://doi.org/10.1073/pnas.1216199109
Zhang J, Li J, Wang X et al (2011) OVP1, a Vacuolar H+-translocating inorganic pyrophosphatase (V-PPase), overexpression improved rice cold tolerance. Plant Physiol Biochem 49(1):33–38. https://doi.org/10.1016/j.plaphy.2010.09.014
Zhang S, Haider I, Kohlen et al (2012) Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol Biol 80(6):571–585. https://doi.org/10.1007/s11103-012-9967-1
Zhang H, Liu Y, Wen F et al (2014) A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J Exp Bot 65(20):5795–5809. https://doi.org/10.1093/jxb/eru313
Zhang A, Yang X, Lu J et al (2021) OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci 308:110903. https://doi.org/10.1016/j.plantsci.2021.110903
Zhang J, Liang Y, Zhang S et al (2022) Global landscape of alternative splicing in maize response to low temperature. J Agric Food Chem 70(50):15715–15725. https://doi.org/10.1021/acs.jafc.2c05969
Zhao J, Wang S, Qin J et al (2019) The lipid transfer protein Os LTPL 159 is involved in cold tolerance at the early seedling stage in rice. Plant Biotechnol J 18(3):756–769. https://doi.org/10.1111/pbi.13243
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumari, A., Sharma, P., Rani, M. et al. Meta-QTL and ortho analysis unravels the genetic architecture and key candidate genes for cold tolerance at seedling stage in rice. Physiol Mol Biol Plants 30, 93–108 (2024). https://doi.org/10.1007/s12298-024-01412-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-024-01412-1