Abstract
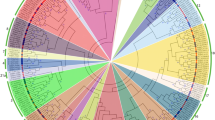
The basic helix-loop-helix (bHLH) is the second-largest TF family in plants that play important roles in plant growth, development, and stress responses. In this study, a total of 100 bHLHs were identified using Hidden Markov Model profiles in the Nicotiana tabacum genome, clustered into 15 major groups (I–XV) based on their conserved domains and phylogenetic relationships. Group VIII genes were found to be the most abundant, with 27 NtbHLH members. The expansion of NtbHLHs in the genome was due to segmental and tandem duplication. The purifying selection was found to have an important role in the evolution of NtHLHs. Subsequent qRT-PCR validation of five selected genes from transcriptome data revealed that NtbHLH3.1, NtbHLH3.2, NtbHLH24, NtbHLH50, and NtbHLH59.2 have higher expressions at 12 and 24 h in comparison to 0 h (control) of chilling stress. The validated results demonstrated that NtbHLH3.2 and NtbHLH24 genes have 3 and fivefold higher expression at 12 h and 2 and threefold higher expression at 24 h than control plant, shows high sensitivity towards chilling stress. Moreover, the co-expression of positively correlated genes of NtbHLH3.2 and NtbHLH24 confirmed their functional significance in chilling stress response. Therefore, suggesting the importance of NtbHLH3.2 and NtbHLH24 genes in exerting control over the chilling stress responses in tobacco.








Similar content being viewed by others
References
Aghdam MS, Sevillano L, Flores FB, Bodbodak S (2013) Heat shock proteins as biochemical markers for postharvest chilling stress in fruits and vegetables. Sci Hortic-Amsterdam 160:54–64
Alessio VM, Cavacana N, Dantas LLD, Lee N, Hotta CT, Imaizumi T, Menossi M (2018) The FBH family of bHLH transcription factors controls ACC synthase expression in sugarcane. J Exp Bot 69:2511–2525
Alptekin B, Langridge P, Budak H (2017) Abiotic stress miRNomes in the triticeae. Funct Integr Genomic 17:145–170
An JP, Wang XF, Zhang XW, Xu HF, Bi SQ, You CX, Hao YJ (2020) An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J 18:337–353
Arnaud N et al (2010) Gibberellins control fruit patterning in arabidopsis thaliana. Genes Dev 24:2127–2132
Arumuganathan K, Martin GB, Telenius H, Tanksley SD, Earle ED (1994) Chromosome 2-specific DNA clones from flow-sorted chromosomes of tomato. Mol Gen Genet 242:551–558
Bailey TL et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202-208
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Bork P, Koonin EV (1996) Protein sequence motifs. Curr Opin Struct Biol 6:366–376
Brownlie P, Ceska TA, Lamers M, Romier C, Stier G, Teo H, Suck D (1997) The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control. Structure 5:509–520
Buck MJ, Atchley WR (2003) Phylogenetic analysis of plant basic helix-loop-helix proteins. J Mol Evol 56:742–750
Burklew CE, Ashlock J, Winfrey WB, Zhang BH (2012) Effects of aluminum oxide nanoparticles on the growth, development, and microRNA expression of tobacco (nicotiana tabacum). PLoS ONE 7(5):e34783
Cao X, Wu Z, Jiang FL, Zhou R, Yang ZE (2014) Identification of chilling stress-responsive tomato microRNAs and their target genes by high-throughput sequencing and degradome analysis. BMC Genomics 15(1):1–6
Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153:1398–1412
Dai XB, Zhuang ZH, Zhao PXC (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46:W49–W54
DeBolt S et al (2009) Mutations in UDP-glucose: sterol glucosyltransferase in arabidopsis cause transparent testa phenotype and suberization defect in seeds. Plant Physiol 151:78–87
Din M, Barozai MYK, Baloch IA (2014) Identification and functional analysis of new conserved microRNAs and their targets in potato (solanum tuberosum L.). Turk J Bot 38:1199
Duek PD, Fankhauser C (2005) bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 10:51–54
Eddy SR (2011) Accelerated profile HMM searches. PLoS Computational Biol 7:e1002195
Edwards KD et al (2017) A reference genome for nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18:448
Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant j: Cell Mol Biol 66:94–116
Ferredamare AR, Pognonec P, Roeder RG, Burley SK (1994) Structure and function of the B/Hlh/Z domain of Usf. Embo J 13:180–189
Finn RD et al (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222–D230
Gao C, Sun JL, Wang CQ, Dong YM, Xiao SH, Wang XJ, Jiao ZG (2017) Genome-wide analysis of basic/helix-loop-helix gene family in peanut and assessment of its roles in pod development. PLoS ONE 12(7):e0181843
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Gaut BS (1998) Molecular clocks and nucleotide substitution rates in higher plants. Evol Biol 30:93–120
Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16:433–442
Gonzalez LE, Keller K, Chan KX, Gessel MM, Thines BC (2017) Transcriptome analysis uncovers Arabidopsis F-BOX STRESS INDUCED 1 as a regulator of jasmonic acid and abscisic acid stress gene expression. BMC Genomics 18(1):1–15. https://doi.org/10.1186/s12864-017-3864-6
Guo AY, Zhu QH, Chen X, Luo JC (2007) [GSDS: a gene structure display server]. Yi chuan = Hereditas 29(8):1023–1026
He XY, Zheng WT, Cao FB, Wu FB (2016) Identification and comparative analysis of the microRNA transcriptome in roots of two contrasting tobacco genotypes in response to cadmium stress. Sci Rep 6(1):1–14. https://doi.org/10.1038/srep32805
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20:735–747
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62:2465–2483
Hu RS, Li ZM, Xiang SP, Li YY, Yi PF, Xiao MQ, Zhang XW (2019) Comparative microRNA profiling reveals the cold response mechanisms in two contrasting tobacco cultivars. Int J Agric Biol 22:757–762
Jones S (2004) An overview of the basic helix-loop-helix proteins. Genome Biol 5:226
Kaur A, Pati PK, Pati AM, Nagpal AK (2017) In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of arabidopsis thaliana and Oryza sativa. PLoS ONE 12(9):e0184523
Kavas M, Baloglu MC, Atabay ES, Ziplar UT, Dasgan HY, Unver T (2016) Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol Genet Genomics 291:129–143
Kim MY, Kang YJ, Lee T, Lee SH (2013) Divergence of flowering-related genes in three legume species. Plant Genome 6(3):plantgenome2013-03. https://doi.org/10.3835/plantgenome2013.03.0008
Koc I, Yuksel I, Caetano-Anolles G (2018) Metabolite-centric reporter pathway and tripartite network analysis of arabidopsis under cold stress. Front Bioeng Biotechnol 6:121
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162
Krzywinski M et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lang TG, Yin KQ, Liu JY, Cao KF, Cannon CH, Du FK (2014) Protein domain analysis of genomic sequence data reveals regulation of LRR related domains in plant transpiration in ficus. PLoS ONE 9(9):e108719
Larkin MA et al (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR (2008) The ups and downs of genome size evolution in polyploid species of nicotiana (Solanaceae). Ann Bot-London 101:805–814
Lescot M et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259
Li AL, Wen Z, Yang K, Wen XP (2019) Conserved miR396b-GRF Regulation Is Involved in Abiotic Stress Responses in Pitaya (Hylocereus polyrhizus). Int J Mol Sci 20(10):2501
Li H et al (2016) High-throughput microRNA and mRNA sequencing reveals that microRNAs may be involved in melatonin-mediated cold tolerance in citrullus lanatus l. Front Plant Sci 7:1231. https://doi.org/10.3389/fpls.2016.01231
Li X et al (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and arabidopsis. Plant Physiol 141:1167–1184
Liang Y et al (2016) Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in brassica napus. Sci Rep 6:24265. https://doi.org/10.1038/srep24265
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in arabidopsis. Plant Cell 10:1391–1406
Liu X, Zhai S, Zhao Y, Sun B, Liu C, Yang A, Zhang J (2013) Overexpression of the phosphatidylinositol synthase gene (ZmPIS) conferring drought stress tolerance by altering membrane lipid composition and increasing ABA synthesis in maize. Plant, Cell Environ 36:1037–1055
Liu X, Zhou Y, Xiao J, Bao F (2018) Effects of chilling on the structure, function and development of chloroplasts. Front Plant Sci 9:1715
Logemann E, Hahlbrock K (2002) Crosstalk among stress responses in plants: Pathogen defense overrides UV protection through an inversely regulated ACE/ACE type of light-responsive gene promoter unit. P Natl Acad Sci USA 99:2428–2432
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic acids research 48(D1):D265–D268. https://doi.org/10.1093/nar/gkz991
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Mao K, Dong QL, Li C, Liu CH, Ma FW (2017) Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front Plant Sci 8:480. https://doi.org/10.3389/fpls.2017.00480
Marchler-Bauer A, Bryant SH (2004) CD-search: protein domain annotations on the fly. Nucleic Acids Res 32:W327–W331
Marin RM, Vanicek J (2011) Efficient use of accessibility in microRNA target prediction. Nucleic Acids Res 39:19–29
Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20:429–440
Murre C, McCaw PS, Baltimore D (1989) A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777–783
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in arabidopsis and grasses. Plant Physiol 149:88–95
Nawaz Z, Kakar KU, Saand MA, Shu QY (2014) Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genomics 15(1):1–8
Nawaz Z, Kakar KU, Ullah R, Yu SZ, Zhang J, Shu QY, Ren XL (2019) Genome-wide identification, evolution and expression analysis of cyclic nucleotide-gated channels in tobacco (Nicotiana tabacum L.). Genomics 111:142–158
Niu X, Guan Y, Chen S, Li H (2017) Genome-wide analysis of basic helix-loop-helix (bHLH) transcription factors in brachypodium distachyon. BMC Genomics 18:619
Ogo Y, Itai R, Nakanishi H, Kobayashi T, Takahashi M, Mori S, Nishizawa N (2007) The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant Cell Physiol 48:S44–S44
Petrussa E, Braidot E, Zancani M, Peresson C, Bertolini A, Patui S, Vianello A (2013) Plant flavonoids-biosynthesis, transport and involvement in stress responses. Int J Mol Sci 14:14950–14973
Pires N, Dolan L (2010) Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol 27:862–874
Pires ND, Dolan L (2012) Morphological evolution in land plants: new designs with old genes. Philos T R Soc B 367:508–518
Qi TC, Huang H, Song SS, Xie DX (2015) Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in arabidopsis. Plant Cell 27:1620–1633
Sah SK, Reddy KR, Li JX (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7:571. https://doi.org/10.3389/fpls.2016.00571
Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ (2017) TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. P Natl Acad Sci USA 114:E897–E903
Shannon P et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Shi Y, Phan H, Liu Y, Cao S, Zhang Z, Chu C, Schlappi MR (2020) Glycosyltransferase OsUGT90A1 helps protect the plasma membrane during chilling stress in rice. J Exp Bot 71:2723–2739
Shimano H (2007) SREBP-1c and TFE3, energy transcription factors that regulate hepatic insulin signaling. J Mol Med 85:437–444
Shimizu T et al (1997) Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. Embo J 16:4689–4697
Shiu SH, Bleecker AB (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in arabidopsis. Plant Physiol 132:530–543
Siddiqi KS, Husen A (2019) Plant response to jasmonates: current developments and their role in changing environment. Bull Natl Res Cent 43:1–11
Sierro N et al (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5(1):1–9. https://doi.org/10.1038/ncomms4833
Song XM, Huang ZN, Duan WK, Ren J, Liu TK, Li Y, Hou XL (2014) Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp pekinensis). Mol Genet Genomics 289:77–91
Song Y et al (2017) High-throughput sequencing of highbush blueberry transcriptome and analysis of basic helix-loop-helix transcription factors. J Integr Agr 16:591–604
Sun H, Fan HJ, Ling HQ (2015) Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16(1):1–2
Sun JH, Sun YH, Ahmed RI, Ren AY, Xie MM (2019) Research progress on plant RING-finger proteins. Genes-Basel 10(12):973
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612
Swamy PM, Smith BN (1999) Role of abscisic acid in plant stress tolerance. Curr Sci India 76:1220–1227
Szecsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M (2006) BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. Embo J 25:3912–3920
Tao X, Wang MX, Dai Y, Wang Y, Fan YF, Mao P, Ma XR (2017) Identification and expression profile of CYPome in perennial ryegrass and tall fescue in response to temperature stress. Front Plant Sci 8:1519. https://doi.org/10.3389/fpls.2017.01519
Thomashow MF (1999) Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Phys 50:571–599
Tokuhisa J, Wu JR, Miquel M, Xin ZU, Browse J (1997) Investigating the role of lipid metabolism in chilling and freezing tolerance. Plant Cold Hardiness 1997:153–169
Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770
Trapnell C et al (2014) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks (vol 7, pg 562, 2012). Nat Protoc 9:2513–2513
Vermaas JV et al (2019) Passive membrane transport of lignin-related compounds. Proc Natl Acad Sci 116:23117–23123
Wang CY (1987) Changes of polyamines and ethylene in cucumber seedlings in response to chilling stress. Physiol Plantarum 69:253–257
Wang LN et al (2014) Genome-wide identification of WRKY family genes and their response to cold stress in vitis vinifera. Bmc Plant Biol 14(1):1–4
Wang TT, Ye CX, Wang M, Chu GM (2017) Identification of cold-stress responsive proteins in anabasis aphylla seedlings via the iTRAQ proteomics technique. J Plant Interact 12:505–519
Wang YP et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40(7):e49
Wei H, Dhanaraj AL, Arora R, Rowland LJ, Fu Y, Sun L (2006) Identification of cold acclimation-responsive rhododendron genes for lipid metabolism, membrane transport and lignin biosynthesis: importance of moderately abundant ESTs in genomic studies. Plant, Cell Environ 29:558–570
Witkos TM, Koscianska E, Krzyzosiak WJ (2011) Practical aspects of microRNA target prediction. Curr Mol Med 11:93–109
Wu HC, Bulgakov VP, Jinn TL (2018) Pectin methylesterases: cell wall remodeling proteins are required for plant response to heat stress. Front Plant Sci 9:1612. https://doi.org/10.3389/fpls.2018.01612
Yang LJ, Yang DB, Yan XJ, Cui L, Wang ZY, Yuan HZ (2016) The role of gibberellins in improving the resistance of tebuconazole-coated maize seeds to chilling stress by microencapsulation. Sci Rep 6(1):1–12. https://doi.org/10.1038/srep35447
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins: structure. Funct, Bioinform 64:643–651
Yu GH, Jiang LL, Ma XF, Xu ZS, Liu MM, Shan SG, Cheng XG (2014) A Soybean C2H2-type zinc finger gene GmZF1 enhanced cold tolerance in transgenic arabidopsis. PLoS ONE 9(10):e109399
Zhang C et al (2018) Genome-wide analysis of basic helix-loop-helix superfamily members in peach. PLoS ONE 13(4):e0195974
Zhang X, Luo HM, Xu ZC, Zhu YJ, Ji AJ, Song JY, Chen SL (2015) Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci Rep 5(1):1–10. https://doi.org/10.1038/srep11244
Zhao Y et al (2019) Functional characterization of a liverworts bHLH transcription factor involved in the regulation of bisbibenzyls and flavonoids biosynthesis. Bmc Plant Biol 19(1):1–13. https://doi.org/10.1186/s12870-019-2109-z
Zheng H et al (2019) Genome-wide identification and analysis of the cytochrome B5 protein family in Chinese cabbage (Brassica rapa L. ssp. Pekinensis). Int J Genomics 2019:2102317
Zhou XF, Wang GD, Sutoh K, Zhu JK, Zhang WX (2008) Identification of cold-inducible microRNAs in plants by transcriptome analysis. Bba-Gene Regul Mech 1779:780–788
Acknowledgements
We thank Dr. Sanchita and Dr. Yogita Deshmukh for correcting the manuscript. The authors acknowledge HPC facility, CSIR-4PI, Bangaluru. This work was supported by a research grant (OLP 0104) from Council of Scientific and Industrial Research (CSIR), India. NB acknowledges UGC SRF fellowship. The manuscript number is provided by the institute is CSIR-NBRI_MS/2020/12/04.
Author information
Authors and Affiliations
Contributions
NB carried out the bioinformatics analysis, design, and drafted the manuscript. PP performed quantitative expression analysis. DC and SKB participated in supervision of the study. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bano, N., Patel, P., Chakrabarty, D. et al. Genome-wide identification, phylogeny, and expression analysis of the bHLH gene family in tobacco (Nicotiana tabacum). Physiol Mol Biol Plants 27, 1747–1764 (2021). https://doi.org/10.1007/s12298-021-01042-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-01042-x