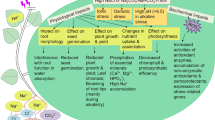
Abstract
Rhizosphere bacteria, whether phytopathogenic or phytobeneficial, are thought to be perceived by the plant as a threat. Plant Growth-Promoting Rhizobacteria (PGPR), such as many strains of the Azospirillum genus known as the main phytostimulator of cereals, cooperate with host plants and favorably affect their growth and health. An earlier study of rice root transcriptome, undertaken with two rice cultivars and two Azospirillum strains, revealed a strain-dependent response during the rice-Azospirillum association and showed that only a few genes, including some implicated in plant defense, were commonly regulated in all tested conditions. Here, a set of genes was selected from previous studies and their expression was monitored by qRT-PCR in rice roots inoculated with ten PGPR strains isolated from various plants and belonging to various genera (Azospirillum, Herbaspirillum, Paraburkholderia). A common expression pattern was highlighted for four genes that are proposed to be markers of the rice-PGPR interaction: two genes involved in diterpenoid phytoalexin biosynthesis (OsDXS3 and OsDTC2) and one coding for an uncharacterized protein (Os02g0582900) were significantly induced by PGPR whereas one defense-related gene encoding a pathogenesis-related protein (PR1b, Os01g0382000) was significantly repressed. Interestingly, exposure to a rice bacterial pathogen also triggered the expression of OsDXS3 while the expression of Os02g0582900 and PR1b was down-regulated, suggesting that these genes might play a key role in rice-bacteria interactions. Integration of these results with previous data led us to propose that the jasmonic acid signaling pathway might be triggered in rice roots upon inoculation with PGPR.


Similar content being viewed by others
Availability of data and material
All used data are reported in the manuscript.
Code availability
Not applicable.
Abbreviations
- IAA:
-
Indole acetic acid
- ISR:
-
Induced systemic resistance
- JA:
-
Jasmonic acid
- MAMP:
-
Microbe-associated molecular patterns
- PGPR:
-
Plant Growth-Promoting Rhizobacteria
References
Ait Barka E, Gognies S, Nowak J, Audran J-C, Belarbi A (2002) Inhibitory effect of endophyte bacteria on Botrytis cinerea and its influence to promote the grapevine growth. Biol Control 24:135–142
Akatsuka T, Sekido H, Takeuchi S, Kono Y, Kodama O (1985) Novel phytoalexins (oryzalexins a, b and c) isolated from rice blast leaves infected with Pyricularia oryzae. Agric Biol Chem 49:1689–1701
Baldani VLD, de Alvarez MAB, Baldani JI, Döbereiner J (1986a) Establishment of inoculated Azospirillum spp. in the rhizosphere and in roots of field grown wheat and sorghum. Plant Soil 90:37–40
Baldani JI, Baldani VLD, Seldin L, Dobereiner J (1986b) Characterization of Herbaspirillurn seropedicae gen. nov. sp. nov. a root-associated nitrogen-fixing bacterium. Int J Syst Bacteriol 36:86–93
Blanvillain-Baufumé S, Reschke M, Solé M, Auguy F, Doucoure H, Szurek B, Meynard D, Portefaix M, Cunnac S, Guiderdoni E, Boch J, Koebnik R (2017) Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol J 15:306–317
Breitler J, Campa C, Georget F, Bertrand B, Etienne H (2016) A single-step method for RNA isolation from tropical crops in the field. Sci Rep 6:38368
Buscaill P, Rivas S (2014) Transcriptional control of plant defence responses. Curr Opin Plant Biol 20:35–46
Cartieaux F, Thibaud MC, Zimmerli L, Lessard P, Sarrobert C, David P, Gerbaud A, Robaglia C, Somerville S, Nussaume L (2003) Transcriptome analysis of Arabidopsis colonized by a plant-growth promoting rhizobacterium reveals a general effect on disease resistance. Plant J 36:177–188
Chamam A, Sanguin H, Bellvert F, Meiffren G, Comte G, Wisniewski-Dyé F, Bertrand C, Prigent-Combaret C (2013) Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum-Oryza sativa association. Phytochemistry 87:65–77
Chen C, Ané JM, Zhu H (2008) OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol 180:311–315
Chen X, Miché L, Sachs S, Wang Q, Buschart A, Yang H, Vera Cruz CM, Hurek T, Reinhold-Hurek B (2015) Rice responds to endophytic colonization which is independent of the common symbiotic signaling pathway. New Phytol 208:531–543
Compant S, Reiter B, Sessitsch A, Nowak J, Clément C, Barka EA (2005) Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl Environ Microbiol 71:1685–1693
Devescovi G, Bigirimana J, Degrassi G, Cabrio L, LiPuma JJ, Kim J, Hwang I, Venturi V (2007) Involvement of a quorum-sensing-regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl Environ Microbiol 73:4950–4958
Dobbelaere S, Vanderleyden J, Okon Y (2003) Plant growth-promoting effects of diazotrophs in the rhizosphere. CRC Crit Rev Plant Sci 22:107–149
Drogue B, Sanguin H, Chamam A, Mozar M, Llauro C, Panaud O, Prigent-Combaret C, Picault N, Wisniewski-Dyé F (2014) Plant root transcriptome profiling reveals a strain-dependent response during Azospirillum-rice cooperation. Front Plant Sci 5:1–14
Egamberdieva D, Wirth SJ, Alqarawi AA, Abd-Allah EF, Hashem A (2017) Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front Microbiol 8:2104
Elbeltagy A, Nishioka K, Sato T, Suzuki H, Ye B, Hamada T, Isawa T, Mitsui H, Minamisawa K (2001) Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. Isolated from wild rice species. Appl Environ Microbiol 67:5285–5293
Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, Hirochika H, Imaizumi-Anraku H, Paszkowski U (2008) Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20:2989–3005
Ito Y, Kurata N (2006) Identification and characterization of cytokinin-signalling gene families in rice. Gene 382:57–65
Kang Y, Kim J, Kim S, Kim H, Lim JY, Kim M, Kwak J, Moon JS, Hwang I (2008) Proteomic analysis of the proteins regulated by HrpB from the plant pathogenic bacterium Burkholderia glumae. Proteomics 8:106–121
King E, Wallner A, Rimbault I, Barrachina A, Klonowska A, Moulin L, Czernic P (2019) Monitoring of rice transcriptional responses to contrasted colonizing patterns of phytobeneficial Burkholderia s.l. reveals a temporal shift in JA systemic response. Front Plant Sci 10:1141
Kost T, Stopnisek N, Agnoli K, Eberl L, Weisskopf L (2013) Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front Microbiol 4:421
Kusajima M, Shima S, Fujita M, Minamisawa K, Che FS, Yamakawa H, Nakashita H (2020) Involvement of ethylene signaling in Azospirillum sp. B510-unduced disease resistance in rice. Biosci Biotechnol Biochem 82:1522–1526
Laborda P, Chen X, Wu GC, Wang SY, Lu XF, Ling J, Li KH, Liu FQ (2020) Lysobacter gummosus OH17 induces systemic resistance in Oryza sativa ‘Nipponbare’. Plant Pathol 69:838–848
Léon-Kloosterziel KM, Verhagen BWM, Keurentjes JJB, Vanpelt JA, Rep M, Vanloon LC, Pieterse CMJ (2005) Colonization of the Arabidopsis rhizosphere by fluorescent Pseudomonas spp. activates a root-specific, ethylene-responsive PR-5 gene in the vascular bundle. Plant Mol Biol 57:731–748
Liang X, Chen X, Li C, Fan J, Guo Z (2017) Metabolic and transcriptional alternations for defense by interfering OsWRKY62 and OsWRKY76 transcriptions in rice. Sci Rep 7:1–15
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
MacDonald MJ, D’Cunha GB (2007) A modern view of phenylalanine ammonia lyase. Biochem Cell Biol 85:759
Marcel S, Sawers R, Oakeley E, Angliker H, Paszkowski U (2010) Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22:3177–3187
Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM (2013) Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci 14:7302–7326
McGee JD, Hamer JE, Hodges TK (2001) Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol Plant-Microbe Interact 14:877–886
Mehnaz S, Lazarovits G (2006) Inoculation effects of Pseudomonas putida, Gluconacetobacter azotocaptans, and Azospirillum lipoferum on corn plant growth under greenhouse conditions. Microb Ecol 51:326–335
Mitsuhara I, Iwai T, Seo S, Yanagawa Y, Kawahigasi H, Hirose S, Ohkawa Y, Ohashi Y (2008) Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds. Mol Genet Genom 279:415–427
Narsai R, Ivanova A, Ng S, Whelan J (2010) Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol 10:56
Nelson LM, Knowles R (1978) Effect of oxygen and nitrate on nitrogen fixation and denitrification by Azospirillum brasilensegrown in continuous culture. Can J Microbiol 24:1395–1403
Okada A, Shimizu T, Okada K, Kuzuyama T, Koga J, Shibuya N, Nojiri H, Yamane H (2007) Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol Biol 65:177–187
Olivares FL, James EK, Baldani JI, Döbereiner J (1997) Infection of mottled stripe disease-susceptible and resistant sugar cane varieties by the endophytic diazotroph Herbaspirillum. New Phytol 135:723–727
Ona O, Van Impe J, Prinsen E, Vanderleyden J (2005) Growth and indole-3-acetic acid biosynthesis of Azospirillum brasilense Sp245 is environmentally controlled. FEMS Microbiol Lett 246:125–132
Pankievicz VCS, Camilios-Neto D, Bonato P, Balsanelli E, Tadra-Sfeir MZ, Faoro H, Chubatsu LS, Donatti L, Wajnberg G, Passetti F, Monteiro RA, Pedrosa FO, Souza EM (2016) RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Mol Biol 90:589–603
Paxton JD (1981) Phytoalexins—a working redefinition. J Phytopathol 101:106–109
Pedrosa FO, Monteiro RA, Wassem R, Cruz LM, Ayub RA, Colauto NB, Fernandez MA et al (2011) Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet 7:e1002064
Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R (2008) OSWRKY62 is a negative regulator of basal and XA21 -mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant 1:446–458
Pereg L, de-Bashan LE, Bashan Y (2016) Assessment of affinity and specificity of Azospirillum for plants. Plant Soil 399:389–414
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375
Rekha K, Kumar RM, Ilango K, Rex A, Usha B (2018) Transcriptome profiling of rice roots in early response to Bacillus subtilis (RR4) colonization. Botany 96:749–765
Rondeau M, Esmaeel Q, Crouzet J, Blin P, Gosselin I, Sarazin C, Pernes M, Beaugrand J, Wisniewski-Dyé F, Vial L, Faure D, Clément C, Ait Barka E, Jacquard C, Sanchez L (2019) Biofilm constructing variants of Paraburkholderia phytofirmans PsJN outcompete the wild-type form in free-living and static conditions but not in planta. Appl Environ Microbiol 85:e02670-18
Schloter M, Hartmann A (1998) Endophytic and surface colonization of wheat roots (Triticum aestivum) by different Azospirillum brasilense strains studied with strain-specific monoclonal antibodies. Symbiosis 25:159–179
Sekido H, Akatsuka T (1987) Mode of action of oryzalexin D against Pyricularia oryzae. Agric Biol Chem 51:1967–1971
Sessitsch A, Coenye T, Sturz AV, Vandamme P, Barka EA, Salles JF, Van Elsas JD, Faure D, Reiter B, Glick BR, Wang-Pruski G, Nowak J (2005) Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int J Syst Evol Microbiol 55:1187–1192
Soto MJ, Domínguez-Ferreras A, Pérez-Mendoza D, Sanjuán J, Olivares J (2009) Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol 11:381–388
Spaepen S, Vanderleyden J, Remans R (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31:425–448
Spaepen S, Vanderleyden J, Okon Y (2009) Plant growth-promoting actions of rhizobacteria. In: Van Loon LC (ed) Plant innate immunity, vol 51. Elsevier, Amsterdam, pp 283–320
Spaepen S, Bossuyt S, Engelen K, Marchal K, Vanderleyden J (2014) Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol 201:850–861
Stringlis IA, Proietti S, Hickman R, Van Verk M, Zamioudis C, Pieterse CMJ (2018) Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J 93:166–180
Tarrand JJ, Krieg NR, Döbereiner J (1978) A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol 24:967–980
Thomas-Bauzon D, Weinhard P, Villecourt P, Balandreau J (1982) The spermosphere model. I. Its use in growing, counting, and isolating N 2 -fixing bacteria from the rhizosphere of rice. Can J Microbiol 28:922–928
Toyomasu T, Kagahara T, Okada K, Koga J, Hasegawa M, Mitsuhashi W, Sassa T, Yamane H (2008) Diterpene phytoalexins are biosynthesized in and exuded from the roots of rice seedlings. Biosci Biotechnol Biochem 72:562–567
Trân Vân V, Ngôkê S, Berge O, Hebbar P, Heulin T, Faure D, Bally R (1997) Isolation of Azospirillum lipoferum from the rhizosphere of rice by a new, simple method. Can J Microbiol 43:486–490
Vacheron J, Moënne-Loccoz Y, Dubost A, Gonçalves-Martins M, Muller D, Prigent-Combaret C (2016) Fluorescent Pseudomonas strains with only few plant-beneficial properties are favored in the maize rhizosphere. Front Plant Sci 7:1–13
Valette M, Rey M, Gerin F, Comte G, Wisniewski-Dyé F (2020) A common metabolomic signature is observed upon inoculation of rice roots with various rhizobacteria. J Integr Plant Biol 62:228–246
van de Mortel JE, de Vos RCH, Dekkers E, Pineda A, Guillod L, Bouwmeester K, van Loon JJA, Dicke M, Raaijmakers JM (2012) Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol 160:2173–2188
Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, van Loon LC, Pieterse CMJ (2004) The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant-Microbe Interact 17:895–908
Vial L, Lavire C, Mavingui P, Blaha D, Haurat J, Moënne-Loccoz Y, Bally R, Wisniewski-Dyé F (2006) Phase variation and genomic architecture changes in Azospirillum. J Bacteriol 188:5364–5373
Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058
Wisniewski-Dyé F, Herrera AM, Drogue B, Acosta-Cruz E, Prigent-Combaret C, Lozano L, González V, Barbe V, Rouy Z, Mavingui P, Borland S (2012) Genome sequence of Azospirillum brasilense CBG497 and comparative analyses of Azospirillum core and accessory genomes provide insight into niche adaptation. Genes 3:576–602
Yamane H (2013) Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Biosci Biotechnol Biochem 77:1141–1148
Yang DL, Yang Y, He Z (2013) Roles of plant hormones and their interplay in rice immunity. Mol Plant 6:675–685
Yang J, Duan GH, Li CQ, Liu L, Han GY, Zhang YL, Wang CM (2020) The crosstalks between jasmonic acid and other plant hormone signaling highlight the Involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front Plant Sci 10:1349
Yano K, Satoko Y, Muller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, Asamizu E, Tabata S, Murooka Y, Perry J, Wang TL, Kawaguchi M, Imaizumi-Anraku H, Hayashi M, Parniske M (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci 105:20540–20545
Zaplin ES, Liu Q, Li Z, Butardo VM, Blanchard CL, Rahman S (2013) Production of high oleic rice grains by suppressing the expression of the OsFAD2-1 gene. Funct Plant Biol 40:996–1004
Zhang X, Bao Y, Shan D, Wang Z, Song X, Wang Z, Wang J, He L, Wu L, Zhang Z, Niu D, Jin H, Zhao H (2018) Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol 177:352–368
Acknowledgements
We wish to thank Emmanuel Guiderdoni (AGAP laboratory, CIRAD Montpellier) for gift of Nipponbare seeds and Lisa Sanchez for providing the GFP-tagged P. phytofirmans PsJN strain. We are grateful to Jonathan Gervaix (AME platform of UMR CNRS-5557) for his invaluable help in acetylene reduction assay, to Danis Abrouk (iBio platform of UMR CNRS-5557) and Samuel Barreto for their great help in statistical analyses. The platform “Serre” of FR41 (University Lyon 1) was used to carry out this work. MV receives a fellowship from «Ministère de l’enseignement supérieur et de la recherche».
Funding
This study was supported by reccurent funding from CNRS and University Lyon 1. MV receives a fellowship from « Ministère de l’enseignement supérieur et de la recherche » .
Author information
Authors and Affiliations
Contributions
M.V. conducted experiments and wrote the paper; M.R., J.D. and F.G. conducted experiments; F.W.D. designed experiments and wrote the paper; all authors discussed the data, and read and approved the contents of this manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethics approval
Not applicable.
Consent to participate
All authors agreed to participate.
Consent for publication
All authors approved the content of this manuscript and provided their consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Valette, M., Rey, M., Doré, J. et al. Identification of a small set of genes commonly regulated in rice roots in response to beneficial rhizobacteria. Physiol Mol Biol Plants 26, 2537–2551 (2020). https://doi.org/10.1007/s12298-020-00911-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-020-00911-1