Abstract
Background
Metastatic spread is characterized by considerable heterogeneity in most cancers. With increasing treatment options for patients with metastatic disease, there is a need for insight into metastatic patterns of spread in breast cancer patients using large-scale studies.
Methods
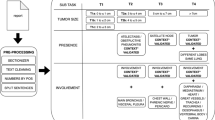
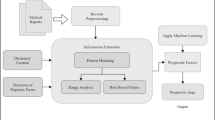
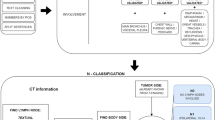
Records of 2622 metastatic breast cancer patients who underwent autopsy (1974–2010) were retrieved from the nationwide Dutch pathology databank (PALGA). Natural language processing (NLP) and manual information extraction (IE) were applied to identify the tumors, patient characteristics, and locations of metastases.
Results
The accuracy (0.90) and recall (0.94) of the NLP model outperformed manual IE (on 132 randomly selected patients). Adenocarcinoma no special type more frequently metastasizes to the lung (55.7%) and liver (51.8%), whereas, invasive lobular carcinoma mostly spread to the bone (54.4%) and liver (43.8%), respectively. Patients with tumor grade III had a higher chance of developing bone metastases (61.6%). In a subgroup of patients, we found that ER+/HER2+ patients were more likely to metastasize to the liver and bone, compared to ER−/HER2+ patients.
Conclusion
This is the first large-scale study that demonstrates that artificial intelligence methods are efficient for IE from Dutch databanks. Different histological subtypes show different frequencies and combinations of metastatic sites which may reflect the underlying biology of metastatic breast cancer.



Similar content being viewed by others
Data availability
The data underlying this article were provided by PALGA and NCR by permission. Data will be shared on reasonable request to the corresponding author with permission from PALGA and NCR.
References
Sørlie T. Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. Eur J Cancer. 2004;40(18):2667–75.
Chia SK, Speers CH, D’yachkova Y, Kang A, Malfair-Taylor S, Barnett J, Coldman A, Gelmon KA, O’Reilly SE, Olivotto IA. The impact of new chemotherapeutic and hormone agents on survival in a population-based cohort of women with metastatic breast cancer. Cancer. 2007;110(5):973–9.
Arciero CA, Guo Y, Jiang R, Behera M, O’Regan R, Peng L, Li X. ER+/HER2+ breast cancer has different metastatic patterns and better survival than ER−/HER2+ breast cancer. Clin Breast Cancer. 2019;19(4):236–45.
Jain S, Fisher C, Smith P, Millis RR, Rubens RD. Patterns of metastatic breast cancer in relation to histological type. Eur J Cancer. 1993;29(15):2155–7.
Cummings MC, Simpson PT, Reid LE, Jayanthan J, Skerman J, Song S, McCart Reed AE, Kutasovic JR, Morey AL, Marquart L, O’Rourke P. Metastatic progression of breast cancer: insights from 50 years of autopsies. J Pathol. 2014;232(1):23–31.
Hugen N, Van de Velde CJ, De Wilt JH, Nagtegaal ID. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. 2014;25(3):651–7.
Verstegen MH, Harker M, van de Water C, van Dieren J, Hugen N, Nagtegaal ID, Rosman C, Van der Post RS. Metastatic pattern in esophageal and gastric cancer: Influenced by site and histology. World J Gastroenterol. 2020;26(39):6037.
Hugen N, Sloot YJ, Netea-Maier RT, van de Water C, Smit JW, Nagtegaal ID, Van Engen-van Grunsven IC. Divergent metastatic patterns between subtypes of thyroid carcinoma results from the nationwide Dutch pathology registry. J Clin Endocrinol Metab. 2020;105(3):e299–306.
Tang R, Ouyang L, Li C, He Y, Griffin M, Taghian A, Smith B, Yala A, Barzilay R, Hughes K. Machine learning to parse breast pathology reports in Chinese. Breast Cancer Res Treat. 2018;169:243–50.
Forsyth AW, Barzilay R, Hughes KS, Lui D, Lorenz KA, Enzinger A, Tulsky JA, Lindvall C. Machine learning methods to extract documentation of breast cancer symptoms from electronic health records. J Pain Symptom Manag. 2018;55(6):1492–9.
Zeng Z, Espino S, Roy A, Li X, Khan SA, Clare SE, Jiang X, Neapolitan R, Luo Y. Using natural language processing and machine learning to identify breast cancer local recurrence. BMC Bioinform. 2018;19(17):65–74.
Van Noord G, Bouma G, Van Eynde F, De Kok D, Van der Linde J, Schuurman I, Sang ET, Vandeghinste V. Large scale syntactic annotation of written Dutch: Lassy. Essential speech and language technology for Dutch: results by the STEVIN programme. 2013;147–64.
Casparie M, Tiebosch AT, Burger G, Blauwgeers H, Van de Pol A, Van Krieken JH, Meijer GA. Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Anal Cell Pathol. 2007;29(1):19–24.
Tan PH, Ellis I, Allison K, Brogi E, Fox SB, Lakhani S, Lazar AJ, Morris EA, Sahin A, Salgado R, Sapino A. The 2019 WHO classification of tumours of the breast. Histopathology. 2020;77:181–5.
Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67(2):93–9.
Medelyan O, Milne D, Legg C, Witten IH. Mining meaning from Wikipedia. Int J Hum Comput Stud. 2009;67(9):716–54.
Bouma G, van Noord G. Increasing return on annotation investment: the automatic construction of a Universal dependency treebank for Dutch. In: Proceedings of the universal dependencies workshop, Gothenburg. 2017. http://aclweb.org/anthology/W17-0403.
van Noord G, et al. Large scale syntactic annotation of written Dutch: Lassy. In: Spyns P, Odijk J, editors., et al., Essential speech and language technology for Dutch. Theory and applications of natural language processing. Berlin: Springer; 2013. https://doi.org/10.1007/978-3-642-30910-6_9.
College of American Pathologists, Gantner GE, Côté RA, Beckett RS. Systematized nomenclature of medicine: coding manual. The College.1979.
Lee YT. Breast carcinoma: pattern of metastasis at autopsy. J Surg Oncol. 1983;23(3):175–80.
Budczies J, von Winterfeld M, Klauschen F, Bockmayr M, Lennerz JK, Denkert C, Wolf T, Warth A, Dietel M, Anagnostopoulos I, Weichert W. The landscape of metastatic progression patterns across major human cancers. Oncotarget. 2015;6(1):570.
Perkins CI, Hotes J, Kohler BA, Howe HL. Association between breast cancer laterality and tumor location, United States, 1994–1998. Cancer Causes Control. 2004;15:637–45.
Cheng SA, Liang LZ, Liang QL, Huang ZY, Peng XX, Hong XC, Luo XB, Yuan GL, Zhang HJ, Jiang L. Breast cancer laterality and molecular subtype likely share a common risk factor. Cancer Manag Res. 2018;29:6549–54.
Abdou Y, Gupta M, Asaoka M, Attwood K, Mateusz O, Gandhi S, Takabe K. Left sided breast cancer is associated with aggressive biology and worse outcomes than right sided breast cancer. Sci Rep. 2022;12(1):13377.
Snoek JAA, Nagtegaal ID, Siesling S, van den Broek E, van Slooten HJ, Hugen N. The impact of standardized structured reporting of pathology reports for breast cancer care. Breast. 2022;66:178–82.
Knijn N, van Erning FN, Overbeek LI, Punt CJ, Lemmens VE, Hugen N, Nagtegaal ID. Limited effect of lymph node status on the metastatic pattern in colorectal cancer. Oncotarget. 2016;7(22):31699.
Banerjee I, Bozkurt S, Caswell-Jin JL, Kurian AW, Rubin DL. Natural language processing approaches to detect the timeline of metastatic recurrence of breast cancer. JCO Clin Cancer Inform. 2019;3:1–2.
De Bruijn LM, Hasman A, Arends JW. Automatic coding of diagnostic reports. Methods Inf Med. 1998;37(03):260–5.
Stanfill MH, Williams M, Fenton SH, Jenders RA, Hersh WR. A systematic literature review of automated clinical coding and classification systems. J Am Med Inform Assoc. 2010;17(6):646–51.
Burger G, Abu-Hanna A, de Keizer N, Cornet R. Natural language processing in pathology: a scoping review. J Clin Pathol. 2016;69(11):949–55.
Nguyen A, Moore D, McCowan I, Courage MJ. Multi-class classification of cancer stages from free-text histology reports using support vector machines. In: 2007 29th Annual international conference of the IEEE Engineering in Medicine and Biology Society. IEEE, (2007);5140–3.
Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42(5):760–72.
Laparra E, Mascio A, Velupillai S, Miller T. A review of recent work in transfer learning and domain adaptation for natural language processing of electronic health records. Yearb Med Inform. 2021;30(01):239–44.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
Conceptualization, MO, NH and IN; Data curation, FK, AS, QV, MO and IN; Funding acquisition, MO and IN; Investigation, FK, AS, MO, NH and IN; Methodology, FK, AS, MO and IN; Project administration, FK, MO and IN; Resources, AS, QV and IN; Supervision, MO and IN; Visualization, FK and IN; Writing—original draft, FK; Writing—review and editing, FK, AS, QV, MO, NH, and IN.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Institutional review board statement
Not applicable.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Kazemzadeh, F., Snoek, J.A.A., Voorham, Q.J. et al. Association of metastatic pattern in breast cancer with tumor and patient-specific factors: a nationwide autopsy study using artificial intelligence. Breast Cancer 31, 263–271 (2024). https://doi.org/10.1007/s12282-023-01534-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-023-01534-6