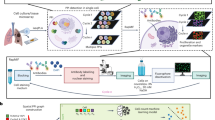
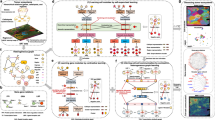
Abstract
Purpose
Current bulk molecular assays fail to capture spatial signaling activities in cancers, limiting our understanding of drug resistance mechanisms. We developed a graph-based super-resolution protein-protein interaction (GSR-PPI) technique to spatially resolve single-cell signaling networks and evaluate whether higher resolution microscopy enhances the biological study of PPIs using deep learning classification models.
Methods
Single-cell spatial proximity ligation assays (PLA, ≤ 9 PPI pairs) were conducted on EGFR mutant (EGFRm) PC9 and HCC827 cells (>10,000 cells) treated with 100 nM Osimertinib. Multiplexed PPI images were obtained using wide-field and super-resolution microscopy (Zeiss Airyscan, SRRF). Graph-based deep learning models analyzed subcellular protein interactions to classify drug treatment states and test GSR-PPI on clinical tissue samples. GSR-PPI triangulated PPI nodes into 3D relationships, predicting drug treatment labels. Biological discriminative ability (BDA) was evaluated using accuracy, AUC, and F1 scores. The method was also applied to 3D spatial proteomic molecular pixelation (PixelGen) data from T cells.
Results
GSR-PPI outperformed baseline models in predicting drug responses from multiplexed PPI imaging in EGFRm cells. Super-resolution data significantly improved accuracy over localized wide-field imaging. GSR-PPI classified drug treatment states in cancer cells and human lung tissues, with performance improving as imaging resolution increased. It differentiated single and combination drug therapies in HCC827 cells and human tissues. Additionally, GSR-PPI accurately distinguished T-cell stimulation states, identifying key nodes such as CD44, CD45, and CD54.
Conclusion
The GSR-PPI framework provides valuable insights into spatial protein interactions and drug responses, enhancing the study of signaling biology and drug resistance.









Similar content being viewed by others
Data Availability
The data that support this published work are available at https://figshare.com/projects/Signaling_Project_PLA/195958. Relevant code can be found at https://github.com/coskunlab/GSR-PPI/tree/main.
References
Fredriksson, S., M. Gullberg, J. Jarvius, C. Olsson, K. Pietras, S. M. Gústafsdóttir, A. Östman, and U. Landegren. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20(5):473–477, 2002. https://doi.org/10.1038/nbt0502-473.
Gridelli, C., A. Rossi, D. P. Carbone, J. Guarize, N. Karachaliou, T. Mok, F. Petrella, L. Spaggiari, and R. Rosell. Non-small-cell lung cancer. Nat. Rev. Disease Primers. 1(1):1–16, 2015. https://doi.org/10.1038/nrdp.2015.9.
Kobayashi, S., T. J. Boggon, T. Dayaram, P. A. Jänne, O. Kocher, M. Meyerson, B. E. Johnson, M. J. Eck, D. G. Tenen, and B. Halmos. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib. New Engl. J. Med. 352(8):786–792, 2005. https://doi.org/10.1056/NEJMoa044238.
Levantini, E., G. Maroni, M. Del Re, and D. G. Tenen. EGFR signaling pathway as therapeutic target in human cancers. Semin. Cancer Biol. Target. Cell. Signal. Pathways. 85:253–275, 2022. https://doi.org/10.1016/j.semcancer.2022.04.002.
Gazdar, A. F. Personalized medicine and inhibition of EGFR signaling in lung cancer. New Engl. J. Med. 361(10):1018–1020, 2009. https://doi.org/10.1056/NEJMe0905763.
Sharma, S. V., D. W. Bell, J. Settleman, and D. A. Haber. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer. 7(3):169–181, 2007. https://doi.org/10.1038/nrc2088.
Liu, X., P. Wang, C. Zhang, and Z. Ma. Epidermal growth factor receptor (EGFR): a rising star in the era of precision medicine of lung cancer. Oncotarget. 8(30):50209–50220, 2017. https://doi.org/10.18632/oncotarget.16854.
Delaney, C., S. Frank, and R. Stephanie Huang. Pharmacogenomics of EGFR-targeted therapies in non–small cell lung cancer: EGFR and beyond. Chin. J. Cancer. 34(3):7, 2015. https://doi.org/10.1186/s40880-015-0007-9.
Uribe, M. L., I. Marrocco, and Y. Yarden. EGFR in cancer: signaling mechanisms, drugs, and acquired resistance. Cancers. 13(11):2748, 2021. https://doi.org/10.3390/cancers13112748.
Fu, K., F. Xie, F. Wang, and L. Fu. Therapeutic strategies for EGFR-mutated non-small cell lung cancer patients with osimertinib resistance. J. Hematol. Oncol. 15(1):173, 2022. https://doi.org/10.1186/s13045-022-01391-4.
Kashima, Y., D. Shibahara, A. Suzuki, K. Muto, I. S. Kobayashi, D. Plotnick, H. Udagawa, H. Izumi, Y. Shibata, K. Tanaka, M. Fujii, A. Ohashi, M. Seki, K. Goto, K. Tsuchihara, Y. Suzuki, and S. S. Kobayashi. Single-cell analyses reveal diverse mechanisms of resistance to EGFR tyrosine kinase inhibitors in lung cancer. Cancer Res. 81(18):4835–4848, 2021. https://doi.org/10.1158/0008-5472.CAN-20-2811.
Leonetti, A., M. Verzè, R. Minari, F. Perrone, L. Gnetti, P. Bordi, M. Pluchino, R. Nizzoli, C. Azzoni, L. Bottarelli, C. A. M. Lagrasta, G. Mazzaschi, S. Buti, D. Gasparro, A. Cosenza, L. Ferri, M. Majori, M. De Filippo, L. Ampollini, S. La Monica, R. Alfieri, E. M. Silini, and M. Tiseo. Resistance to osimertinib in advanced EGFR-mutated NSCLC: a prospective study of molecular genotyping on tissue and liquid biopsies. Br. J. Cancer. 130(1):135–142, 2024. https://doi.org/10.1038/s41416-023-02475-9.
Wang, X. F., B. Liang, D. X. Zeng, W. Lei, C. Chen, Y. B. Chen, J. A. Huang, N. Gu, and Y. H. Zhu. The roles of MASPIN expression and subcellular localization in non-small cell lung cancer. Biosci. Rep. 40(5):BSR20200743, 2020. https://doi.org/10.1042/BSR20200743.
Drexler, R., R. Fahy, M. Küchler, K. C. Wagner, T. Reese, M. Ehmke, B. Feyerabend, M. Kleine, and K. J. Oldhafer. Association of subcellular localization of TEAD transcription factors with outcome and progression in pancreatic ductal adenocarcinoma. Pancreatology. 21(1):170–179, 2021. https://doi.org/10.1016/j.pan.2020.12.003.
Yi, Y., P. Li, Y. Huang, D. Chen, S. Fan, J. Wang, M. Yang, S. Zeng, J. Deng, X. Lv, K. Luo, Z. He, and H. Liu. P21-activated kinase 2-mediated β-catenin signaling promotes cancer stemness and osimertinib resistance in EGFR-mutant non-small-cell lung cancer. Oncogene. 41(37):4318–4329, 2022. https://doi.org/10.1038/s41388-022-02438-z.
Jiajia, G., W. Yao, P. Shi, G. Zhang, T. K. Owonikoko, S. S. Ramalingam, and S.-Y. Sun. MEK or ERK inhibition effectively abrogates emergence of acquired osimertinib resistance in the treatment of EGFR-mutant lung cancers. Cancer. 126(16):3788–3799, 2020. https://doi.org/10.1002/cncr.32996.
Jacobsen, K., J. Bertran-Alamillo, M. A. Molina, C. Teixidó, N. Karachaliou, M. H. Pedersen, J. Castellví, M. Garzón, C. Codony-Servat, J. Codony-Servat, and A. Giménez-Capitán. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat. Commun. 8(1):410, 2017. https://doi.org/10.1038/s41467-017-00450-6.
Kurppa, K. J., Y. Liu, C. To, T. Zhang, M. Fan, A. Vajdi, E. H. Knelson, Y. Xie, K. Lim, P. Cejas, and A. Portell. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 37(1):104-122.e12, 2020. https://doi.org/10.1016/j.ccell.2019.12.006.
Fu, M., Y. Hu, T. Lan, K. L. Guan, T. Luo, and M. Luo. The Hippo signalling pathway and its implications in human health and diseases. Signal Trans. Target. Ther. 7(1):376, 2022. https://doi.org/10.1038/s41392-022-01191-9.
Arasada, R. R., K. Shilo, T. Yamada, J. Zhang, S. Yano, R. Ghanem, W. Wang, S. Takeuchi, K. Fukuda, N. Katakami, and K. Tomii. Notch3-dependent β-catenin signaling mediates EGFR TKI drug persistence in EGFR mutant NSCLC. Nat. Commun. 9:3198, 2018. https://doi.org/10.1038/s41467-018-05626-2.
Casás-Selves, M., J. Kim, Z. Zhang, B. A. Helfrich, D. Gao, C. C. Porter, H. A. Scarborough, P. A. Bunn Jr., D. C. Chan, A. C. Tan, and J. DeGregori. Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res. 72(16):4154–4164, 2012. https://doi.org/10.1158/0008-5472.CAN-11-2848.
Nakayama, S., N. Sng, J. Carretero, R. Welner, Y. Hayashi, M. Yamamoto, A. J. Tan, N. Yamaguchi, H. Yasuda, D. Li, K. Soejima, R. A. Soo, D. B. Costa, K.-K. Wong, and S. S. Kobayashi. β-catenin contributes to lung tumor development induced by EGFR mutations. Cancer Res. 74(20):5891–5902, 2014. https://doi.org/10.1158/0008-5472.CAN-14-0184.
Chang, H. A., R. Z. Ou Yang, J. M. Su, T. M. H. Nguyen, J. M. Sung, M. J. Tang, and W. T. Chiu. YAP nuclear translocation induced by HIF-1α prevents DNA damage under hypoxic conditions. Cell Death Discov. 9(1):1–14, 2023. https://doi.org/10.1038/s41420-023-01687-5.
Hsu, P.-C., C.-T. Yang, D. M. Jablons, and L. You. The crosstalk between Src and hippo/YAP signaling pathways in non-small cell lung cancer (NSCLC). Cancers. 12(6):1361, 2020. https://doi.org/10.3390/cancers12061361.
Jiang, L., J. Li, C. Zhang, Y. Shang, and J. Lin. YAP-mediated crosstalk between the Wnt and Hippo signaling pathways (Review). Mol. Med. Rep. 22(5):4101–4106, 2020. https://doi.org/10.3892/mmr.2020.11529.
Liu, Z., D. Xing, S. Qian Peter, Y. Zhu, J. Zhang, X. Kong, B. Xue, S. Wang, H. Sun, Y. Tao, and Y. Sun. Super-resolution imaging and tracking of protein–protein interactions in sub-diffraction cellular space. Nat. Commun. 5(1):4443, 2014. https://doi.org/10.1038/ncomms5443.
Oi, C., Z. Gidden, L. Holyoake, O. Kantelberg, S. Mochrie, M. H. Horrocks, and L. Regan. LIVE-PAINT allows super-resolution microscopy inside living cells using reversible peptide-protein interactions. Commun. Biol. 3(1):1–10, 2020. https://doi.org/10.1038/s42003-020-01188-6.
Martens, K. J. A., B. Turkowyd, and U. Endesfelder, Raw data to results: a hands-on introduction and overview of computational analysis for single-molecule localization microscopy, Front. Bioinform. 1, 2022
Sauer, M. Localization microscopy coming of age: from concepts to biological impact. J. Cell Sci. 126(16):3505–3513, 2013. https://doi.org/10.1242/jcs.123612.
Martens, K. J. A., A. N. Bader, S. Baas, B. Rieger, and J. Hohlbein. Phasor based single-molecule localization microscopy in 3D (pSMLM-3D): an algorithm for MHz localization rates using standard CPUs. J. Chem. Phys. 148(12):123311, 2017. https://doi.org/10.1063/1.5005899.
Khater, I. M., I. R. Nabi, and G. Hamarneh. A review of super-resolution single-molecule localization microscopy cluster analysis and quantification methods. Patterns. 1(3):100038, 2020. https://doi.org/10.1016/j.patter.2020.100038.
Wolter, S., T. Holm, S. van de Linde, and M. Sauer, Data analysis for single-molecule localization microscopy. In: Super-Resolution Microscopy Techniques in the Neurosciences. Edited by E. F. Fornasiero, and S. O. Rizzoli, Humana Press: Totowa, 2014, pp. 113–132. Online at https://doi.org/10.1007/978-1-62703-983-3_6 (accessed February 10, 2024).
Brenner, B., C. Sun, F. M. Raymo, and H. F. Zhang. Spectroscopic single-molecule localization microscopy: applications and prospective. Nano Converg. 10(1):14, 2023. https://doi.org/10.1186/s40580-023-00363-9.
Stringer, C., T. Wang, M. Michaelos, and M. Pachitariu. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods. 18(1):100–106, 2021. https://doi.org/10.1038/s41592-020-01018-x.
Dinas, S., and H. J. Martínez, Delaunay triangulation, In: Encyclopedia of Computer Graphics and Games. Edited by N. Lee, Springer: Cham, 2020, pp. 1–6. Online at https://doi.org/10.1007/978-3-319-08234-9_393-1. Accessed 29 January 2024.
Kipf, T. N., and M. Welling, Semi-supervised classification with graph convolutional networks, Preprint, arXiv, February 22, 2017. https://doi.org/10.48550/arXiv.1609.02907.
Veličković P., G. Cucurull, A. Casanova, A. Romero, P. Liò, and Y. Bengio, Graph attention networks, Preprint, arXiv, February 4, 2018. https://doi.org/10.48550/arXiv.1710.10903.
Brody, S., U. Alon, and E. Yahav, How attentive are graph attention networks?, Preprint, arXiv, January 31, 2022. https://doi.org/10.48550/arXiv.2105.14491.
Morris, C., M. Ritzert, M. Fey, W. L. Hamilton, J. E. Lenssen, G. Rattan, and M. Grohe, Weisfeiler and leman go neural: higher-order graph neural networks, Preprint, arXiv, November 30, 2021. https://doi.org/10.48550/arXiv.1810.02244.
Li, Y., D. Tarlow, M. Brockschmidt, and R. Zemel, Gated graph sequence neural networks, Preprint, arXiv, September 22, 2017. https://doi.org/10.48550/arXiv.1511.05493.
Hayes, T. K., E. Aquilanti, N. S. Persky, X. Yang, E. E. Kim, L. Brenan, A. B. Goodale, D. Alan, T. Sharpe, R. E. Shue, and L. Westlake. Comprehensive mutational scanning of EGFR reveals TKI sensitivities of extracellular domain mutants. Nat. Commun. 15(1):2742, 2024. https://doi.org/10.1038/s41467-024-45594-4.
Girard, L., S. Zochbauer-Muller, A. K. Virmani, A. F. Gazdar, and J. D. Minna. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 60(17):4894–4906, 2000.
Engelman, J. A., K. Zejnullahu, T. Mitsudomi, Y. Song, C. Hyland, J. O. Park, N. Lindeman, C. M. Gale, X. Zhao, J. Christensen, and T. Kosaka. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 316(5827):1039–1043, 2007. https://doi.org/10.1126/science.1141478.
Park, M. Y., M. H. Jung, E. Y. Eo, S. Kim, S. H. Lee, Y. J. Lee, J. S. Park, Y. J. Cho, J. H. Chung, C. H. Kim, and H. I. Yoon. Generation of lung cancer cell lines harboring EGFR T790M mutation by CRISPR/Cas9-mediated genome editing. Oncotarget. 8(22):36331–36338, 2017. https://doi.org/10.18632/oncotarget.16752.
de Jager, V. D., J. A. Stigt, M. Niemantsverdriet, A. Ter Elst, and A. J. van der Wekken. Osimertinib and palbociclib in an EGFR-mutated NSCLC with primary CDK4 amplification after progression under osimertinib. NPJ Precis. Oncol. 8(1):1–4, 2024. https://doi.org/10.1038/s41698-024-00607-9.
Rao, G., M. Pierobon, I. K. Kim, W. H. Hsu, J. Deng, Y. W. Moon, E. F. Petricoin, Y. W. Zhang, Y. Wang, and G. Giaccone. Inhibition of AKT1 signaling promotes invasion and metastasis of non-small cell lung cancer cells with K-RAS or EGFR mutations. Sci. Rep. 7(1):7066, 2017. https://doi.org/10.1038/s41598-017-06128-9.
Terp, M. G., K. Jacobsen, M. A. Molina, N. Karachaliou, H. C. Beck, J. Bertran-Alamillo, A. Giménez-Capitán, A. F. Cardona, R. Rosell, and H. J. Ditzel. Combined FGFR and Akt pathway inhibition abrogates growth of FGFR1 overexpressing EGFR-TKI-resistant NSCLC cells. NPJ Precis. Oncol. 5(1):65, 2021. https://doi.org/10.1038/s41698-021-00208-w.
Yuan, B.-Z., A. M. Jefferson, L. Millecchia, N. C. Popescu, and S. H. Reynolds. Morphological changes and nuclear translocation of DLC1 tumor suppressor protein precede apoptosis in human non-small cell lung carcinoma cells. Exp. Cell Res. 313(18):3868–3880, 2007. https://doi.org/10.1016/j.yexcr.2007.08.009.
Malhotra, J., B. Ryan, M. Patel, N. Chan, Y. Guo, J. Aisner, S. K. Jabbour, and S. Pine. Clinical outcomes and immune phenotypes associated with STK11 co-occurring mutations in non-small cell lung cancer. J. Thorac. Disease. 14(6):1772, 2022. https://doi.org/10.21037/jtd-21-1377.
Sumbly V., and I. Landry, Unraveling the role of STK11/LKB1 in non-small cell lung cancer. Cureus. 14(1): e21078. https://doi.org/10.7759/cureus.21078.
Application note: multiplex mode for the LSM 9 series with Airyscan 2: fast and gentle confocal super-resolution in large volumes.
Henriques, R., M. Lelek, E. F. Fornasiero, F. Valtorta, C. Zimmer, and M. M. Mhlanga. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods. 7(5):339–340, 2010. https://doi.org/10.1038/nmeth0510-339.
Jiang, T., and Y. Qiu. Interaction between Src and a C-terminal proline-rich motif of Akt is required for Akt activation. J.Biol. Chem. 278(18):15789–15793, 2003. https://doi.org/10.1074/jbc.M212525200.
Tsytlonok, M., H. Sanabria, Y. Wang, S. Felekyan, K. Hemmen, A. H. Phillips, M. K. Yun, M. B. Waddell, C. G. Park, S. Vaithiyalingam, and L. Iconaru. Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation. Nat. Commun. 10(1):1676, 2019. https://doi.org/10.1038/s41467-019-09446-w.
Song, B., H. Ge, C. Pu, and N. Li. GLP2-GLP2R signal affects the viability and EGFR-TKIs sensitivity of PC9 and HCC827 cells. BMC Pulm. Med. 22(1):36, 2022. https://doi.org/10.1186/s12890-021-01800-3.
Pang, W., Y. Li, W. Guo, and H. Shen. Cyclin E: a potential treatment target to reverse cancer chemoresistance by regulating the cell cycle. Am. J. Trans. Res. 12(9):5170, 2020.
Josefsberg Ben-Yehoshua, L., K. Beider, A. Shimoni, O. Ostrovsky, M. Samookh, A. Peled, and A. Nagler. Characterization of cyclin E expression in multiple myeloma and its functional role in seliciclib-induced apoptotic cell death. Plos One. 7(4):e33856, 2012. https://doi.org/10.1371/journal.pone.0033856.
Petty, W. J., W. R. Voelzke, J. J. Urbanic, V. A. Varela, L. L. Waller, C. B. Swift, R. M. Graham, V. A. Memoli, and K. H. Dragnev. High cyclin D3 expression confers erlotinib resistance in aerodigestive tract cancer. Lung Cancer. 74(3):384–391, 2011. https://doi.org/10.1016/j.lungcan.2011.04.004.
Raina, D., Y. Uchida, A. Kharbanda, H. Rajabi, G. Panchamoorthy, C. Jin, S. Kharbanda, M. Scaltriti, J. Baselga, and D. Kufe. Targeting the MUC1-C oncoprotein downregulates HER2 activation and abrogates trastuzumab resistance in breast cancer cells. Oncogene. 33(26):3422–3431, 2014. https://doi.org/10.1038/onc.2013.308.
Liu, J. J., J. Y. Ho, H. W. Lee, M. W. Baik, O. Kim, Y. J. Choi, and S. Y. Hur. Inhibition of phosphatidylinositol 3-kinase (PI3K) signaling synergistically potentiates antitumor efficacy of paclitaxel and overcomes paclitaxel-mediated resistance in cervical cancer. Int. J. Mol. Sci. 20(14):3383, 2019. https://doi.org/10.3390/ijms20143383.
Xie, C., Y. Han, L. Fu, Q. Li, X. Qiu, and E. Wang. Overexpression of CARMA3 is associated with advanced tumor stage, cell cycle progression, and cisplatin resistance in human epithelial ovarian cancer. Tumor Biol. 35(8):7957–7964, 2014. https://doi.org/10.1007/s13277-014-2070-2.
Fagundes, R., and L. K. Teixeira, Cyclin E/CDK2: DNA replication, replication stress and genomic instability, Front. Cell Dev. Biol. 9, 2021
Gustafsson, N., S. Culley, G. Ashdown, D. M. Owen, P. M. Pereira, and R. Henriques. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat. Commun. 7(1):12471, 2016. https://doi.org/10.1038/ncomms12471.
Lee, G., C. Wong, A. Cho, J. J. West, A. J. Crawford, G. C. Russo, B. R. Si, J. Kim, L. Hoffner, C. Jang, and M. Jung. E-cadherin induces serine synthesis to support progression and metastasis of breast cancer. Cancer Res. 2024. https://doi.org/10.1158/0008-5472.CAN-23-3082.
Kase, S., K. Sugio, K. Yamazaki, T. Okamoto, T. Yano, and K. Sugimachi. Expression of E-cadherin and β-catenin in human non-small cell lung cancer and the clinical significance. Clinic. Cancer Res. 6(12):4789–4796, 2000.
Karlsson, F., T. Kallas, D. Thiagarajan, M. Karlsson, M. Schweitzer, J. F. Navarro, L. Leijonancker, S. Geny, E. Pettersson, J. Rhomberg-Kauert, and L. Larsson. Molecular pixelation: spatial proteomics of single cells by sequencing. Nat. Methods. 21(6):1044–1052, 2024. https://doi.org/10.1038/s41592-024-02268-9.
Kennedy-Darling, J., O. Shang, C. Hempel, N. Jhaveri, N. Nikulina, O. Braubach, B. B. Cheikh, and J. Yuan. Chapter 4 - Highly multiplexed spatial protein data using CODEX technology. In: Revealing Unchartered Biology with Single Intact Cells, Edited by W. J. Fantl, Academic Press, 2024, pp. 93–118. Online at https://www.sciencedirect.com/science/article/pii/B9780128222096000011. Accessed 16 July 2024
Zhao, Y., Z. X. Li, Y. J. Zhu, J. Fu, X. F. Zhao, Y. N. Zhang, S. Wang, J. M. Wu, K. T. Wang, R. Wu, and C. J. Sui. Single-cell transcriptome analysis uncovers intratumoral heterogeneity and underlying mechanisms for drug resistance in hepatobiliary tumor organoids. Adv. Sci. 8(11):2003897, 2021. https://doi.org/10.1002/advs.202003897.
Aissa, A. F., A. B. Islam, M. M. Ariss, C. C. Go, A. E. Rader, R. D. Conrardy, A. M. Gajda, C. Rubio-Perez, K. Valyi-Nagy, M. Pasquinelli, and L. E. Feldman. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat. Commun. 12(1):1628, 2021. https://doi.org/10.1038/s41467-021-21884-z.
Liu, P., J. Liu, J. Liu, and X. Yu. Investigating the mechanisms of drug resistance and prognosis in ovarian cancer using single-cell RNA sequencing and bulk RNA sequencing. Aging (Albany NY). 16(5):4736–4758, 2024. https://doi.org/10.18632/aging.205628.
Kim, K. T., H. W. Lee, H. O. Lee, S. C. Kim, Y. J. Seo, W. Chung, H. H. Eum, D. H. Nam, J. Kim, K. M. Joo, and W. Y. Park. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genom. Biol. 16(1):1–15, 2015. https://doi.org/10.1186/s13059-015-0692-3.
Eyler, C. E., H. Matsunaga, V. Hovestadt, S. J. Vantine, P. van Galen, and B. E. Bernstein. Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genom. Biol. 21(1):1–21, 2020. https://doi.org/10.1186/s13059-020-02085-1.
Gene-specific correlation of RNA and protein levels in human cells and tissues, Mol. Syst. Biol. 12(10): 883, 2016 https://doi.org/10.15252/msb.20167144.
Gry, M., R. Rimini, S. Strömberg, A. Asplund, F. Pontén, M. Uhlén, and P. Nilsson. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genom. 10(1):1–14, 2009. https://doi.org/10.1186/1471-2164-10-365.
Maier, T., M. Güell, and L. Serrano. Correlation of mRNA and protein in complex biological samples. FEBS let. Syst. Biol.—Nobel Symp. 146. 583(24):3966–3973, 2009. https://doi.org/10.1016/j.febslet.2009.10.036.
Alam, M. S. Proximity ligation assay (PLA)Proximity ligation assay (PLA). In: Immunohistochemistry and Immunocytochemistry: Methods and Protocols, Edited by L. D. Valle, New York: Springer, 2022, pp. 191–201. Online at https://doi.org/10.1007/978-1-0716-1948-3_13. Accessed 15 February 2024.
Cheng, Y., R. K. Burrack, and Q. Li. Spatially resolved and highly multiplexed protein and RNA In situ detection by combining CODEX With RNAscope in situ hybridization. J. Histochem. Cytochem. 70(8):571–581, 2022. https://doi.org/10.1369/00221554221114174.
Merritt, C. R., G. T. Ong, S. E. Church, K. Barker, P. Danaher, G. Geiss, M. Hoang, J. Jung, Y. Liang, J. McKay-Fleisch, and K. Nguyen. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38(5):586–599, 2020. https://doi.org/10.1038/s41587-020-0472-9.
Vistain, L., H. Van Phan, B. Keisham, C. Jordi, M. Chen, S. T. Reddy, and S. Tay. Quantification of extracellular proteins, protein complexes and mRNAs in single cells by proximity sequencing. Nat. Methods. 19(12):1578–1589, 2022. https://doi.org/10.1038/s41592-022-01684-z.
Acknowledgments
AFC holds a Career Award at the Scientific Interface from Burroughs Wellcome Fund and a Bernie-Marcus Early-Career Professorship. A. F. C. was supported by start-up funds from the Georgia Institute of Technology and Emory University. Research reported in this publication was supported by Lung Spore and the National Cancer Institute of the National Institutes of Health under Award Number P50CA217691 from the Career Enhancement Program and R35GM151028. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Research reported in this publication was supported in part by the Cancer Tissue and Pathology Shared Resource and the Data and Technology Applications Shared Resource of Winship Cancer Institute of Emory University and NIH/NCI under award number P30CA138292. The research was also supported by 1R33CA291197.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Coskun, Cai, and Hu declare a patent application related to the spatial signaling interactomics assay (US Provisional 63/399,427 and US Application No 18/452,178). Nicholas Zhang, Mingshuang Wang, Frank Schneider, and Shi-Yong Sun declare no competing interests.
Ethical Approval
The study did not directly involve animal or human subjects. The use of human specimens was approved by the Institutional Review Board of Emory University (IRB00098377).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the CMBE 2024 Young Innovators special issue.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, N., Cai, S., Wang, M. et al. Graph-Based Spatial Proximity of Super-Resolved Protein–Protein Interactions Predicts Cancer Drug Responses in Single Cells. Cel. Mol. Bioeng. 17, 467–490 (2024). https://doi.org/10.1007/s12195-024-00822-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-024-00822-1
Keywords
Profiles
- Ahmet F. Coskun View author profile