Abstract
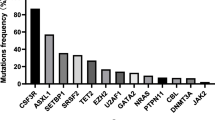
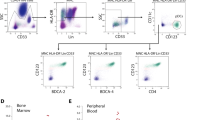
Disease relapse is a major cause of treatment failure after allogeneic hematopoietic cell transplantation (HCT) and the mechanisms of relapse remain unclear. We encountered a 58-year-old man with chronic myelomonocytic leukemia (CMML) that relapsed after haploidentical HCT from his daughter. Peripheral blood samples collected at HCT and at relapse were analyzed, and CD14+/CD16− monocytes that typically accumulate in CMML were isolated by flow cytometry. Whole-exome sequencing of the monocytes revealed 8 common mutations in CMML at HCT. In addition, a PHF6 nonsense mutation not detected at HCT was detected at relapse. RNA sequencing could not detect changes in expression of HLA or immune-checkpoint molecules, which are important mechanisms of immune evasion. However, gene set enrichment analysis (GSEA) revealed that a TNF-α signaling pathway was downregulated at relapse. Ubiquitination of histone H2B at lysine residue 120 (H2BK120ub) at relapse was significantly decreased at the protein level, indicating that PHF6 loss might downregulate a TNF-α signaling pathway by reduction of H2BK120ub. This case illustrates that PHF6 loss contributes to a competitive advantage for the clone under stress conditions and leads to relapse after HCT.


Similar content being viewed by others
References
Zeiser R, Vago L. Mechanisms of immune escape after allogeneic hematopoietic cell transplantation. Blood. 2019;133:1290–7.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Valent P, Orazi A, Savona MR, Patnaik MM, Onida F, van de Loosdrecht AA, et al. Proposed diagnostic criteria for classical chronic myelomonocytic leukemia (CMML), CMML variants and pre-CMML conditions. Haematologica. 2019;104:1935–49.
Savona MR, Malcovati L, Komrokji R, Tiu RV, Mughal TI, Orazi A, et al. An international consortium proposal of uniform response criteria for myelodysplastic/myeloproliferative neoplasms (MDS/MPN) in adults. Blood. 2015;125:1857–65.
Selimoglu-Buet D, Wagner-Ballon O, Saada V, Bardet V, Itzykson R, Bencheikh L, et al. Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia. Blood. 2015;125:3618–26.
Patnaik MM, Tefferi A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6: e393.
Patel BJ, Przychodzen B, Thota S, Radivoyevitch T, Visconte V, Kuzmanovic T, et al. Genomic determinants of chronic myelomonocytic leukemia. Leukemia. 2017;31:2815–23.
Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31:2428–36.
Gagelmann N, Badbaran A, Beelen DW, Salit RB, Stolzel F, Rautenberg C, et al. A prognostic score including mutation profile and clinical features for patients with CMML undergoing stem cell transplantation. Blood Adv. 2021;5:1760–9.
Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJ, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med. 2018;379:2330–41.
Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci G, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med. 2019;25:603–11.
Miyagi S, Sroczynska P, Kato Y, Nakajima-Takagi Y, Oshima M, Rizq O, et al. The chromatin-binding protein Phf6 restricts the self-renewal of hematopoietic stem cells. Blood. 2019;133:2495–506.
Pronk CJ, Veiby OP, Bryder D, Jacobsen SE. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. J Exp Med. 2011;208:1563–70.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730–5.
Ramirez-Herrick AM, Mullican SE, Sheehan AM, Conneely OM. Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms in mice. Blood. 2011;117:2681–90.
Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H, Wong LL. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One. 2013;8: e55761.
Palomo L, Malinverni R, Cabezon M, Xicoy B, Arnan M, Coll R, et al. DNA methylation profile in chronic myelomonocytic leukemia associates with distinct clinical, biological and genetic features. Epigenetics. 2018;13:8–18.
Meacham CE, Lawton LN, Soto-Feliciano YM, Pritchard JR, Joughin BA, Ehrenberger T, et al. A genome-scale in vivo loss-of-function screen identifies Phf6 as a lineage-specific regulator of leukemia cell growth. Genes Dev. 2015;29:483–8.
Oh S, Boo K, Kim J, Baek SA, Jeon Y, You J, et al. The chromatin-binding protein PHF6 functions as an E3 ubiquitin ligase of H2BK120 via H2BK12Ac recognition for activation of trophectodermal genes. Nucleic Acids Res. 2020;48:9037–52.
Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26.
Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373:1550–61.
Franzini A, Pomicter AD, Yan D, Khorashad JS, Tantravahi SK, Than H, et al. The transcriptome of CMML monocytes is highly inflammatory and reflects leukemia-specific and age-related alterations. Blood Adv. 2019;3:2949–61.
Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25:130–4.
Quek L, Ferguson P, Metzner M, Ahmed I, Kennedy A, Garnett C, et al. Mutational analysis of disease relapse in patients allografted for acute myeloid leukemia. Blood Adv. 2016;1:193–204.
Acknowledgements
We would like to thank all the clinicians at our center who helped obtain data for this study. Y. Akahoshi is a Research Fellow of Japan Society of the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Contributions
YA designed the study, performed the experiments, collected data, analyzed data, and wrote the manuscript. HN advised on methods and reviewed the manuscript. MKu, KK, YN, MKa, JT, SKa, NY, YM, KY, SM, AG, AT, MT, S-iK, and SKa collected important data. YK reviewed the manuscript, revised the manuscript, and organized this project.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest for this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Akahoshi, Y., Nakasone, H., Kusuda, M. et al. Emergence of clone with PHF6 nonsense mutation in chronic myelomonocytic leukemia at relapse after allogeneic HCT. Int J Hematol 115, 748–752 (2022). https://doi.org/10.1007/s12185-021-03284-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-021-03284-7