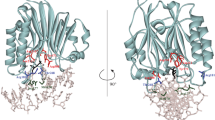
Abstract
Apurinic/apyrimidinic endonuclease 1 (APE1 or Ref-1) is the major enzyme in mammals for processing abasic sites in DNA. These cytotoxic and mutagenic lesions arise via spontaneous rupture of the base-sugar bond or the removal of damaged bases by a DNA glycosylase. APE1 cleaves the DNA backbone 5′ to an abasic site, giving a 3′-OH primer for repair synthesis, and mediates other key repair activities. The DNA repair functions are essential for embryogenesis and cell viability. APE1-deficient cells are hypersensitive to DNA-damaging agents, and APE1 is considered an attractive target for inhibitors that could potentially enhance the efficacy of some anti-cancer agents. To enable an important new method for studying the structure, dynamics, catalytic mechanism, and inhibition of APE1, we assigned the chemical shifts (backbone and 13Cβ) of APE1 residues 39-318. We also report a protocol for refolding APE1, which was essential for achieving complete exchange of backbone amide sites for the perdeuterated protein.


Similar content being viewed by others
References
Beernink PT, Segelke BW, Hadi MZ, Erzberger JP, Wilson DM 3rd, Rupp B (2001) Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: implications for the catalytic mechanism. J Mol Biol 307:1023–1034
Bhakat KK, Mantha AK, Mitra S (2009) Transcriptional regulatory functions of mammalian ap-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal 11:621–637
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Fishel ML, Kelley MR (2007) The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Asp Med 28:375–395
Fitzgerald ME, Drohat AC (2008) Coordinating the initial steps of base excision repair: apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. J Biol Chem 283:32680–32690
Goddard TD, Kneller DG. SPARKY 3. University of California, San Francisco
Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, Hickson ID, Freemont PS (1997) The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J 16:6548–6558
Jung YS, Zweckstetter M (2004) Mars—robust automatic backbone assignment of proteins. J Biomol NMR 30:11–23
McNeill DR, Lam W, DeWeese TL, Cheng YC, Wilson DM 3rd (2009) Impairment of APE1 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol Cancer Res 7:897–906
Mol CD, Izumi T, Mitra S, Tainer JA (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403:451–456
Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K (1999) TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc 121:844–848
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS plus: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
Willis MS, Hogan JK, Prabhakar P, Liu X, Tsai K, Wei Y, Fox T (2005) Investigation of protein refolding using a fractional factorial screen: a study of reagent effects and interactions. Protein Sci 14:1818–1826
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01-GM72711 to A.C.D.) and the University of Maryland Marlene and Stewart Greenebaum Cancer Center. B.M. is supported by a Chemistry-Biology Interface (CBI) Training grant from the NIH, T32-GM066706. The NMR spectrometers used in these studies were purchased, in part, with funds from shared instrumentation grants from the NIH (S10-RR10441; S10-RR15741; S10-RR16812; S10-RR23447) and the NSF (DBI 0115795). We thank Mike Morgan for producing the expression plasmid for APE1ΔN38.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Manvilla, B.A., Varney, K.M. & Drohat, A.C. Chemical shift assignments for human apurinic/apyrimidinic endonuclease 1. Biomol NMR Assign 4, 5–8 (2010). https://doi.org/10.1007/s12104-009-9196-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-009-9196-y