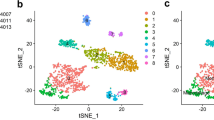
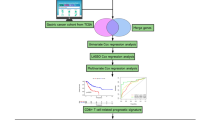
Abstract
Background
Neoadjuvant immunotherapy has evolved as an effective option to treat non-small cell lung cancer (NSCLC). B cells play essential roles in the immune system as well as cancer progression. However, the repertoire of B cells and its association with clinical outcomes remains unclear in NSCLC patients receiving neoadjuvant immunotherapy.
Methods
Single-cell RNA sequencing (scRNA-seq) and bulk RNA sequencing data for LUAD samples were accessed from the TCGA and GEO databases. LUAD-related B cell marker genes were confirmed based on comprehensive analysis of scRNA-seq data. We then constructed the B cell marker gene signature (BCMGS) and validated it. In addition, we evaluated the association of BCGMS with tumor immune microenvironment (TIME) characteristics. Furthermore, we validated the efficacy of BCGMS in a cohort of NSCLC patients receiving neoadjuvant immunotherapy.
Results
A BCMGS was constructed based on the TCGA cohort and further validated in three independent GSE cohorts. In addition, the BCMGS was proven to be significantly associated with TIME characteristics. Moreover, a relatively higher risk score indicated poor clinical outcomes and a worse immune response among NSCLC patients receiving neoadjuvant immunotherapy.
Conclusions
We constructed an 18-gene prognostic signature derived from B cell marker genes based on scRNA-seq data, which had the potential to predict the prognosis and immune response of NSCLC patients receiving neoadjuvant immunotherapy.





Similar content being viewed by others
Data availability
The datasets generated or analyzed during the current study are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and TCGA database (https://portal.gdc.cancer.gov/).
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics. CA Cancer J Clin. 2022;72(1):7–33.
Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–54.
Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. NCCN Guidelines® insights: non-small cell lung cancer, Version 2. J Nat Comp Cancer Network JNCCN. 2023;21(4):340–50.
Leighl NB, Hellmann MD, Hui R, Carcereny E, Felip E, Ahn MJ, et al. Pembrolizumab in patients with advanced non-small-cell lung cancer (KEYNOTE-001): 3-year results from an open-label, phase 1 study. Lancet Respir Med. 2019;7(4):347–57.
Chaft JE, Oezkan F, Kris MG, Bunn PA, Wistuba II, Kwiatkowski DJ, et al. Neoadjuvant atezolizumab for resectable non-small cell lung cancer: an open-label, single-arm phase II trial. Nat Med. 2022;28(10):2155–61.
Cascone T, William WN Jr, Weissferdt A, Leung CH, Lin HY, Pataer A, et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in operable non-small cell lung cancer: the phase 2 randomized NEOSTAR trial. Nat Med. 2021;27(3):504–14.
Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–22.
Baba Y, Saito Y, Kotetsu Y. Heterogeneous subsets of B-lineage regulatory cells (Breg cells). Int Immunol. 2020;32(3):155–62.
Downs-Canner SM, Meier J, Vincent BG, Serody JS. B cell function in the tumor microenvironment. Annu Rev Immunol. 2022;40:169–93.
Chen H, Ye F, Guo G. Revolutionizing immunology with single-cell RNA sequencing. Cell Mol Immunol. 2019;16(3):242–9.
Zhou R, Zhou J, Muhuitijiang B, Zeng X, Tan W. Construction and experimental validation of a B cell-related gene signature to predict the prognosis and immunotherapeutic sensitivity in bladder cancer. Aging. 2023;15(12):5355–80.
Zhang F, Guo W, Zhou B, Wang S, Li N, Qiu B, et al. Three-year follow-up of neoadjuvant programmed cell death protein-1 Inhibitor (Sintilimab) in NSCLC. J Thoracic Oncol Off Publ Int Assoc Study Lung Cancer. 2022;17(7):909–20.
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The immune landscape of cancer. Immunity. 2018;48(4):812-830.e14.
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–62.
Li H, Ayer LM, Lytton J, Deans JP. Store-operated cation entry mediated by CD20 in membrane rafts. J Biol Chem. 2003;278(43):42427–34.
Duan H, Chen B, Wang W, Luo H. Identification of GNG7 as a novel biomarker and potential therapeutic target for gastric cancer via bioinformatic analysis and in vitro experiments. Aging. 2023;15(5):1445–74.
Sun X, Sun Z, Zhu Z, Guan H, Zhang J, Zhang Y, et al. Clinicopathological significance and prognostic value of lactate dehydrogenase A expression in gastric cancer patients. PLoS ONE. 2014;9(3): e91068.
Wang YY, Li L, Zhao ZS, Wang HJ. Clinical utility of measuring expression levels of KAP1, TIMP1 and STC2 in peripheral blood of patients with gastric cancer. World J Surg Oncol. 2013;11:81.
Song G, Xu S, Zhang H, Wang Y, Xiao C, Jiang T, et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer Res CR. 2016;35(1):148.
Miyamoto K, Iwadate M, Yanagisawa Y, Ito E, Imai J, Yamamoto M, et al. Cathepsin L is highly expressed in gastrointestinal stromal tumors. Int J Oncol. 2011;39(5):1109–15.
Pinto JA, Rolfo C, Raez LE, Prado A, Araujo JM, Bravo L, et al. In silico evaluation of DNA damage inducible transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies. Sci Rep. 2017;7(1):1526.
Zeng Q, Liu J, Cao P, Li J, Liu X, Fan X, et al. Inhibition of REDD1 sensitizes bladder urothelial carcinoma to paclitaxel by inhibiting autophagy. Clin Cancer Res Off J Am Assoc Cancer Res. 2018;24(2):445–59.
Jin HO, Hong SE, Kim JY, Kim MR, Chang YH, Hong YJ, et al. Induction of HSP27 and HSP70 by constitutive overexpression of Redd1 confers resistance of lung cancer cells to ionizing radiation. Oncol Rep. 2019;41(5):3119–26.
Yan J, Huang QY, Huang YJ, Wang CS, Liu PX. SPATS2 is positively activated by long noncoding RNA SNHG5 via regulating DNMT3a expression to promote hepatocellular carcinoma progression. PLoS ONE. 2022;17(1): e0262262.
Urabe F, Kosaka N, Sawa Y, Ito K, Kimura T, Egawa S, et al. The miR-1908/SRM regulatory axis contributes to extracellular vesicle secretion in prostate cancer. Cancer Sci. 2020;111(9):3258–67.
Ding Z, Joy M, Bhargava R, Gunsaulus M, Lakshman N, Miron-Mendoza M, et al. Profilin-1 downregulation has contrasting effects on early vs late steps of breast cancer metastasis. Oncogene. 2014;33(16):2065–74.
Wang Z, Shi Z, Zhang L, Zhang H, Zhang Y. Profilin 1, negatively regulated by microRNA-19a-3p, serves as a tumor suppressor in human hepatocellular carcinoma. Pathol Res Pract. 2019;215(3):499–505.
Karamchandani JR, Gabril MY, Ibrahim R, Scorilas A, Filter E, Finelli A, et al. Profilin-1 expression is associated with high grade and stage and decreased disease-free survival in renal cell carcinoma. Hum Pathol. 2015;46(5):673–80.
Wang M, Xiao X, Zeng F, Xie F, Fan Y, Huang C, et al. Common and differentially expressed long noncoding RNAs for the characterization of high and low grade bladder cancer. Gene. 2016;592(1):78–85.
Capello M, Ferri-Borgogno S, Cappello P, Novelli F. α-Enolase: a promising therapeutic and diagnostic tumor target. FEBS J. 2011;278(7):1064–74.
Mitchell CB, Black B, Sun F, et al. Tropomyosin Tpm 2.1 loss induces glioblastoma spreading in soft brain-like environments. J Neuro-Oncol. 2019;141(2):303–13.
Wu Z, Ge L, Ma L, Lu M, Song Y, Deng S, et al. TPM2 attenuates progression of prostate cancer by blocking PDLIM7-mediated nuclear translocation of YAP1. Cell Biosci. 2023;13(1):39.
Wang B, Zhong JL, Li HZ, Wu B, Sun DF, Jiang N, et al. Diagnostic and therapeutic values of PMEPA1 and its correlation with tumor immunity in pan-cancer. Life Sci. 2021;277: 119452.
Wolff DW, Xie Y, Deng C, Gatalica Z, Yang M, Wang B, et al. Epigenetic repression of regulator of G-protein signaling 2 promotes androgen-independent prostate cancer cell growth. Int J Cancer. 2012;130(7):1521–31.
Ying L, Lin J, Qiu F, Cao M, Chen H, Liu Z, et al. Epigenetic repression of regulator of G-protein signaling 2 by ubiquitin-like with PHD and ring-finger domain 1 promotes bladder cancer progression. FEBS J. 2015;282(1):174–82.
Lee SM, Shin H, Jang SW, Shim JJ, Song IS, Son KN, et al. PLP2/A4 interacts with CCR1 and stimulates migration of CCR1-expressing HOS cells. Biochem Biophys Res Commun. 2004;324(2):768–72.
Longo A, Librizzi M, Luparello C. Effect of transfection with PLP2 antisense oligonucleotides on gene expression of cadmium-treated MDA-MB231 breast cancer cells. Anal Bioanal Chem. 2013;405(6):1893–901.
Wang L, Wang L, Zhang H, Lu J, Zhang Z, Wu H, et al. AREG mediates the epithelial-msesenchymal transition in pancreatic cancer cells via the EGFR/ERK/NF-κB signalling pathway. Oncol Rep. 2020;43(5):1558–68.
Ivagnès A, Messaoudene M, Stoll G, Routy B, Fluckiger A, Yamazaki T, et al. TNFR2/BIRC3-TRAF1 signaling pathway as a novel NK cell immune checkpoint in cancer. Oncoimmunology. 2018;7(12): e1386826.
Barnes TA, Amir E. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br J Cancer. 2017;117(4):451–60.
Liu C, Zhao W, Xie J, Lin H, Hu X, Li C, et al. Development and validation of a radiomics-based nomogram for predicting a major pathological response to neoadjuvant immunochemotherapy for patients with potentially resectable non-small cell lung cancer. Front Immunol. 2023;14:1115291.
Funding
This work was supported by the National Key R&D Program of China (2021YFC2500900), National Natural Science Foundation of China (82273129), Medical and Health Science-Technology Innovation Project of the Chinese Academy of Medical Sciences (2021-I2M-1-015), Central Health Research Key Projects (2022ZD17), and Research Project of the Institute (LC2019L01).
Author information
Authors and Affiliations
Contributions
LY (Liu Yang) analyzed the public data curation and was a major contributor in writing the manuscript. BFL collected the genomic data and performed the initial analysis of the data. BGY participated in coming up with the idea regarding the neoadjuvant immunotherapy and performed book retrieval for group discussion. HQL performed the initial review of the manuscript. LY (Li Yuan) plotted the figures and wrote the figure legends. CXW performed data interpretation and provided the methodological supportation. ZBL provided the methodological supportation and participated in writing the original draft. GSG was the corresponding author of the manuscript and provided funding acquisition. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Y., Bie, F., Bai, G. et al. Prognostic model based on B cell marker genes for NSCLC patients under neoadjuvant immunotherapy by integrated analysis of single-cell and bulk RNA-sequencing data. Clin Transl Oncol (2024). https://doi.org/10.1007/s12094-024-03428-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12094-024-03428-1