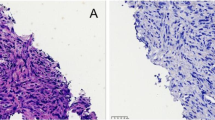
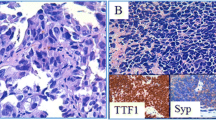
Abstract
Purpose
Aberrant activation of MET as a result of exon 14-skipping (METex14) mutations or gene amplification is an oncogenic mechanism in non-small cell lung carcinoma (NSCLC) and a potential therapeutic target. The purpose of this study was to characterize MET alterations in a cohort of NSCLC patients treated with surgery.
Methods and patients
157 NSCLCs of various histopathologies, including pulmonary sarcomatoid carcinomas (PSC), were tested for MET alterations. METex14 mutations, MET copy number alterations and the levels of MET protein were determined by Sanger sequencing, fluorescence in situ hybridization and immunohistochemistry, respectively. Concurrent alterations of other important cancer genes and immunostaining of the downstream effector, phopho-S6, were also determined.
Results
METex14 mutations and MET amplification were detected in seven tumors. MET genetic alterations were found predominantly in the lung adenocarcinoma (ADC) and PSC histopathologies. High levels of MET protein were found in most MET-amplified tumors, but not in all METex14-mutated tumors. Strong phopho-S6 staining was observed in about half of the MET-activated tumors. One tumor with METex14 exhibited concurrent ERBB2 amplification.
Conclusions
MET activation, by either METex14 mutations or amplification, is characteristic of a subset of early stage NSCLCs and may coexist with ERBB2 amplification. This may have potential therapeutic implications. The presence of METex14 mutations was associated with low levels of MET protein, which may limit the use of total MET immunostaining as a marker for preselecting patients for MET-targeted therapies.


Similar content being viewed by others
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86.
Conway K, Price P, Harding KG, Jiang WG. The molecular and clinical impact of hepatocyte growth factor, its receptor, activators, and inhibitors in wound healing. Wound Repair Regen. 2006;14:2–10.
Furlan A, Kherrouche Z, Montagne R, Copin M-C, Tulasne D. Thirty years of research on met receptor to move a biomarker from bench to bedside. Cancer Res. 2014;74:6737–44.
Okuda K, Sasaki H, Yukiue H, Yano M, Fujii Y. Met gene copy number predicts the prognosis for completely resected non-small cell lung cancer. Cancer Sci. 2008;99:2280–5.
Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–9.
Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–7.
Comprehensive molecular profiling of lung adenocarcinoma. Cancer Genome Atlas Research Network. Nature. 2014;511:543–50.
Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–16.
Onozato R, Kosaka T, Kuwano H, Sekido Y, Yatabe Y, Mitsudomi T. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol. 2009;4:5–11.
Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–9.
Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, et al. MET exon 14 mutations in non–small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol. 2016;34:721–30.
Scagliotti GV, Novello S, Schiller JH, Hirsh V, Sequist LV, Soria JC, et al. Rationale and design of MARQUEE: a phase III, randomized, double-blind study of tivantinib plus erlotinib versus placebo plus erlotinib in previously treated patients with locally advanced or metastatic, nonsquamous, non-small-cell lung cancer. Clin Lung Cancer. 2012;13:391–5.
Spigel DR, Edelman MJ, O’Byrne K, Paz-Ares L, Shames DS, Yu W, et al. Onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIb or IV NSCLC: results from the pivotal phase III randomized, multicenter, placebo-controlled METLung (OAM4971 g) global trial. J Clin Oncol. 2014;32:5 (Suppl; abstr 8000).
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–60.
Conde E, Angulo B, Tang M, Morente M, Torres-Lanzas J, Lopez-Encuentra A, et al. Molecular context of the EGFR mutations: evidence for the activation of mTOR/S6 K signaling. Clin Cancer Res. 2006;12:710–7.
Angulo B, Suarez-Gauthier A, Lopez-Rios F, Medina PP, Conde E, Tang M, et al. Expression signatures in lung cancer reveal a profile for EGFR-mutant tumours and identify selective PIK3CA overexpression by gene amplification. J Pathol. 2008;214:347–56.
Pros E, Lantuejoul S, Sanchez-Verde L, Castillo SD, Bonastre E, Suarez-Gauthier A, et al. Determining the profiles and parameters for gene amplification testing of growth factor receptors in lung cancer. Int J Cancer. 2013;133:898–907.
Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, et al. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable met gene mutations. J Clin Oncol. 2016;34:794–802.
Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, et al. characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol. 2016;11:1493–502.
Tong JH, Yeung SF, Chan AWH, Chung LY, Chau SL, Lung RWM, et al. MET Amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin Cancer Res. 2016;22:3048–56.
Seo J-S, Ju YS, Lee W-C, Shin J-Y, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–19.
Hömig-Hölzel C, Savola S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn Mol Pathol. 2012;21:189–206.
Walsh PS, Erlich HA, Higuchi R. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1992;1:241–50.
Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–74.
Tyner JW, Fletcher LB, Wang EQ, Yang WF, Rutenberg-Schoenberg ML, Beadling C, et al. MET receptor sequence variants R970C and T992I lack transforming capacity. Cancer Res. 2010;70:6233–7.
Zheng D, Wang R, Ye T, Yu S, Hu H, Shen X, et al. MET exon 14 skipping defines a unique molecular class of non-small cell lung cancer. Oncotarget. 2016;7:41691–702.
Acknowledgements
This work was supported by several Spanish grants: SAF2014-54571-R, co-funded by FEDER funds/European Regional Development Fund (ERDF)-“a Way to Build Europe” (to M Sanchez-Cespedes); AGAUR, Agency to Grups de recerca consolidats (2014SGR641) (to M Sanchez-Cespedes); Fondo de Investigaciones Sanitarias (PI14/01109 to E. Nadal); and the Fundación Científica Asociación Española Contra el Cáncer-GCB14-2170.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to disclose.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Research involving human participants
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saigi, M., McLeer-Florin, A., Pros, E. et al. Genetic screening and molecular characterization of MET alterations in non-small cell lung cancer. Clin Transl Oncol 20, 881–888 (2018). https://doi.org/10.1007/s12094-017-1799-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-017-1799-7