Abstract
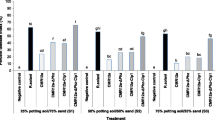
Pseudomonas maltophila PM-4, an antagonist of pathogenic fungi including Rhizoctonia bataticola, R. solani, Fusarium oxysporum and Sclerotinia sclerotiorum associated with root rot of clusterbean (Cyamopsis tetragonoloba) was mutagenized with Tn5. Hyperchitinase producing mutants showing large zone of colloidal chitin dissolution were identified on medium containing calcoflor dye as an indicator. A mutant P-48 producing 137% higher chitinase activity than the parent strain PM-4 was identified. Seed bacterization of clusterbean (Cyamopsis tetragonoloba) with P-48 controlled the root rot upto 40.8% in the presence of conglomerate of all the four fungal pathogens Rhizoctonia bataticola, R. solani, F. oxysporum and Sclerotinia sclerotiorum.
Similar content being viewed by others
References
Singh JV, Rai L & Arora RM (2001) Dynamics of clusterbean production in India. Indian Farming 3:36–38
Lodha S (1998) Effect of inoculum on population dynamics of Macrophomina phaseolina and disease intensity in clusterbean. Indian Phytopathol 57:175–179
Weller D (1988) Biological control of soil borne plant pathogens in the rhizosphere with bacteria. Ann Rev Phytopathol 26:379–407
Lonhienne T, Mavromatis K, Vorgias CE, Buchon L, Gerday C & Bouriotis V (2001) Cloning, sequences, and characterization of two chitinase genes from the Antarctic Arthrobacter sp. strain TAD20: isolation and partial characterization of the enzymes. Appl Environ Microbiol 63:1773–1779
Tsujibo H, Orikoshi H, Baba N, Miyahara M, Katsushiro M, Masahide Y & Yoshihiko I (2002) Identification and characterization of the gene cluster involved in chitin degradation in a marine bacterium, Alteromonas sp. Strain O-7. Appl Environ Microbiol 68:263–270
Gohel V, Chaudhary T, Vyas P & Chhatpar HS (2004) Isolation and identification of marine chitinolytic bacteria and their potential in antifungal biocontrol. Ind J Exp Biol 42:715–720
Nandakumar R, Babu S, Radjacommare R, Raguchander T & Samiyappan R (2002) Pseudomonas fluorescens mediated antifungal activity against Rhizoctonia solani causing sheath blight in rice. Phytopathologia Mediterranea 41:109–119
Raaijmakers JM. & Weller DM (2001) Exploiting genetic diversity of 2, 4 diacetylpholoroglucinol producing Pseudomonas spp.: characterization of superior root —colonizing P. fluorescens strain Q8rl-96. Appl Environ Microbiol 67:2545–2554
Dileep C & Kumar BS (2002) Influence of Metham Sodium on suppression of color rot disease of peanut, in vitro antibiosis, siderophore production and root colonization by fluorescent pseudomonads strain FPO4. Ind J Exp Biol 38:1245–1250
Kremer RJ & Souissi T (2001) Cyanide production by rhizobacteria and potential for suppression of weed seedlings growth. Curr Microbiol 43:182–186
Ton J, Davison S, Van Wees SC, Van Loon LC & Pieterse CM (2001) The arabidopsis ISR locus controlling rhizobacteria-mediated induced systemic resistance is involved in ethylene signaling. Plant Physiol 125:652–661
Kamensky M, Ovadis M, Chet I & Chernin L (2003) Soil borne strain IC14 of Serratia plymuthica with multiple mechanisms of antifungal activity provides biocontrol of Botrytis cinerea and Sclerotinia sclerotiorum diseases. Soil Biol Biochem 35:323–331
Sundheim L, Poplawsky AB & Ellingboe AH (1988) Molecular cloning of two chitinase genes from Serratia marcescens and their expression in Pseudomonas species. Physiol Mol Plant Pathol 33:483–491
Kohli U (2001) Integrated control of root rot pathogen of sunflower by antagonistic bacteria. Ph.D. Thesis, CCS Haryana Agricultural University, Hisar
Zhang Z & Yuen G (2000) The role of chitinase production by Stenotrophomonas maltophila strain C3 in biological control of Bipolaris sorokiniana. Phytopathology 90:384–389
Lingappa Y & Lockwood JL (1961) Chitin media for selective isolation and culture of actinomycetes. Phytopathology 52:317–323
Kambhoj DV, Sharma PK & Kundu, BS (1996) Direct monitoring of Rhizobium sp. (cicer) in nodule and soil using lac Z fusions. Biol Fertil Soils 21:309–313
Simon R, Priefer UB & Puehler A (1983) A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784–791
Vaidya R J, Macmil SLA, Vyas PR & Chhatpar HS (2003) The novel method for isolating chitinolytic bacteria and its application in screening for hyperchitinase producing mutants of Alcaligenes xylosoxydens. Lett Appl Microbiol 36:129–134
Morrissey RF, Dugen EP & Koths JS (1976) Chitinase production by an Arthrobacter sp. lysing cells of Fusarium roseum. Soil Biol Biochem 8:22–28
Schwyn B & Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Annals Biochem 160:47–56
Castric PA (1975) Hydrogen cyanide, a secondary metabolite of Pseudomanas aeruginosa. Can J Microbiol 21:613–618
Ordentlich A, Elad Y & Chet I (1988) The role of chitinase of Serratia marcescens in biocontrol of Sclerotium rolfsii. Phytopathology 78:84–88
Kobayashi DY, Guglielmoni M. & Clarke BB (1995) Isolation of the chitinolytic bacteria Xanthomonas maltophila and Serratia marcescens as biological control agents for summer patch disease of turf grass. Soil Biol Biochem 27:1479–1487
Frankowski J, Berg G & Bahl H (1998) Mechanisms involved in the antifungal activity of the rhizobacterium Serratia plymuthica. IOBC Bulletin 9:45–50
Zhang Z & Yuen G (2000) Effect of culture fluid and preinduction on chitinase production on Bipolaris leaf spot by Stenotrophomonas maltophila. Biocontrol 18:277–286
Garg R, Sharma PK & Kundu, BS (1996) Role of Azospirillum exopolysacchiredes in root colonization of pearl millet (Pennisaetum americana). Ind J Microbiol 36:193–197
Glazebrook J & Walker GC (1990) A novel exopolysaccharide can function in place of calcoflor binding exopolysccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661–672
Watanabe T, Kimura K, Sumiya T, Nikaidou N, Suzuki K, Suzuki M, Taiyoji M., Ferrer S & Regue M (1997) Genetic analysis of the chitinase system of Serratia marcescens 2170. J Bacteriol 179:7111–7117
Techkarnjanarruk S, Pongpattanakitshote S & Goodman AE (1997) Use of promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp strain S9. Appl Environ Microbiol 63:2989–2996
Pal KK, Tilak KV, Saxena AK, Dey R & Singh, CS (2001) Suppression of maize root disease by Macrophomina phaseolina, Fusarium moniliforma and Fusarium graminearum by PGPR. Microbiol Res 100:209–213
Chet I, Ordentlich A, Shapira R & Oppenheim A (1990) Mechanism of biocontrol of soil borne plant pathogens by rhizobacteria. Plant & Soil 129:85–92
Kobayashi DY, Reedy RM, Bick JA & Oudemans PV (2002) Characterization of a chitinase gene from Strenotrophomonas maltophila strain 34S1 and its involvement in biological control. Appl Environ Microbiol 68:1047–1054
Jacobi M, Winkelmann G, Kaiser D, Kempter C, Jung G, Berg G & Bahl H (1996) Maltophilin, a new antifungal compound produced by Stenotrophomonas maltophila R3089. J Antibiotics 49:1104–1109
Dunne C, Crowley JJ, Monne-Loccoz D, Dowling DN, de Bruijn F J & O’Gara F (1997) Biological control of Pythium ultimum by Stenotrophomanas maltophila W81 is mediated by an extracellular proteolytic activity. Microbiology 143:3921–3931
Nakayama T, Homma Y, Hashidoko Y, Mizutani J & Tahara S (1999) Possible role of xanthobactins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping off disease. Appl Environ Microbiol 65:4334–4339
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yadav, E., Pathak, D.V., Sharma, S.K. et al. Isolation and characterization of mutants of Pseudomonas maltophila PM-4 altered in chitinolytic activity and antagonistic activity against root rot pathogens of clusterbean (Cyamopsis tetragonoloba). Indian J Microbiol 47, 64–71 (2007). https://doi.org/10.1007/s12088-007-0012-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-007-0012-z