Abstract
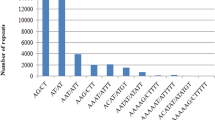
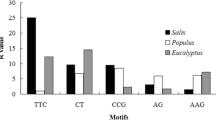
Pineapple (Ananas comosus (L.) Merrill) is the second most important tropical fruit in term of international trade. The availability of whole genomic sequences and expressed sequence tags (ESTs) offers an opportunity to identify and characterize microsatellite or simple sequence repeat (SSR) markers in pineapple. A total of 278,245 SSRs and 41,962 SSRs with an overall density of 728.57 SSRs/Mb and 619.37 SSRs/Mb were mined from genomic and ESTs sequences, respectively. 5′-untranslated regions (5′-UTRs) had the greatest amount of SSRs, 3.6–5.2 fold higher SSR density than other regions. For repeat length, 12 bp was the predominant repeat length in both assembled genome and ESTs. Class I SSRs were underrepresented compared with class II SSRs. For motif length, dinucleotide repeats were the most abundant in genomic sequences, whereas trinucleotides were the most common motif in ESTs. Tri- and hexanucleotides of total SSRs were more prevalent in ESTs than in the whole genome. The SSR frequency decreased dramatically as repeat times increased. AT was the most frequent single motif across the entire genome while AG was the most abundant motif in ESTs. Across six examined plant species, the pineapple genome displayed the highest density, substantially more than the second-place cucumber. Annotation and expression analyses were also conducted for genes containing SSRs. This thorough analysis of SSR markers in pineapple provided valuable information on the frequency and distribution of SSRs in the pineapple genome. This genomic resource will expedite genomic research and pineapple improvement.








Similar content being viewed by others
Abbreviations
- AFLP:
-
amplified fragment length polymorphism
- bp:
-
Base pair
- CAM:
-
Crassulacean Acid Metabolism
- CDS:
-
Coding sequences
- EST:
-
Expressed sequence tag
- FPKM:
-
Number of fragments per kilobase of exon per million fragments mapped
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- kb:
-
Kilo base pairs
- MAS:
-
Marker-assisted selection
- Mb:
-
Mega base pairs
- RAPD:
-
Randomly amplified polymorphic DNA
- RFLP:
-
Restriction fragment length polymorphism
- SSR:
-
Simple sequence repeat
- STR:
-
Short tandem repeat
- UTR:
-
Untranslated regions
References
Aradhya MK, Zee F, Manshardt RM (1994) Isozyme variation in cultivated and wild pineapple. Euphytica 79:87–99
Arumuganathan K, Earle E (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Report 9:229–241
Ayres N, McClung A, Larkin P, Bligh H, Jones C, Park W (1997) Microsatellites and a single-nucleotide polymorphism differentiate apparentamylose classes in an extended pedigree of US rice germ plasm. Theor Appl Genet 94:773–781
Biet E, Sun J-S, Dutreix M (1999) Conserved sequence preference in DNA binding among recombination proteins: an effect of ssDNA secondary structure. Nucleic Acids Res 27:596–600
Biswas MK, Xu Q, Mayer C, Deng X (2014) Genome wide characterization of short tandem repeat markers in sweet Orange (Citrus sinensis). PLoS One 9. doi:10.1371/journal.pone.0104182
Carlier JD, Sousa NH, Santo TE, d’Eeckenbrugge GC, Leitão JM (2012) A genetic map of pineapple (Ananas comosus (L.) merr.) including SCAR, CAPS, SSR and EST-SSR markers. Mol Breed 29:245–260
Castillo A, Budak H, Varshney RK, Dorado G, Graner A, Hernandez P (2008) Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis in Hordeum chilense. BMC Plant Biol 8:97
Cavagnaro PF et al. (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genomics 11:569
Chen H et al. (2015) A high-density SSR genetic map constructed from a F2 population of Gossypium hirsutum and Gossypium darwinii. Gene 574:273–286
de Sousa N, Carlier J, Santo T, Leitão J (2013) An integrated genetic map of pineapple (Ananas comosus (L.) merr.). Sci Hortic-Amsterdam 157:113–118
DeWald M, Moore G, Sherman W (1992) Isozymes in Ananas (pineapple): genetics and usefulness in taxonomy. J Am Soc Hortic Sci 117:491–496
Dresselhaus T, Cordts S, Heuer S, Sauter M, Lörz H, Kranz E (1999) Novel ribosomal genes from maize are differentially expressed in the zygotic and somatic cell cycles. Mol Gen Genet 261:416–427
Duval M, Noyer J, Perrier X, d’Eeckenbrugge C, Hamon P (2001) Molecular diversity in pineapple assessed by RFLP markers. Theor Appl Genet 102:83–90
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5:435–445
Feng S et al. (2013) Development of pineapple microsatellite markers and germplasm genetic diversity analysis. Biomed Res Int 2013:11
Fraser L, Harvey C, Crowhurst R, De Silva H (2004) EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108:1010–1016
Gailing O, Bodénès C, Finkeldey R, Kremer A, Plomion C (2013) Genetic mapping of EST-derived simple sequence repeats (EST-SSRs) to identify QTL for leaf morphological characters in a Quercus robur full-sib family. Tree Genet Genomes 9:1361–1367
Garza JC, Slatkin M, Freimer NB (1995) Microsatellite allele frequencies in humans and chimpanzees, with implications for constraints on allele size. Mol Biol Evol 12:594–603
Grabherr MG et al. (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–652
Gur-Arie R, Cohen CJ, Eitan Y, Shelef L, Hallerman EM, Kashi Y (2000) Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res 10:62–71
Harding RM, Boyce A, Clegg J (1992) The evolution of tandemly repetitive DNA: recombination rules. Genetics 132:847–859
Huo N et al. (2008) The nuclear genome of Brachypodium distachyon: analysis of BAC end sequences. Funct Integr Genomics 8:135–147
Innan H, Terauchi R, Miyashita NT (1997) Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics 146:1441–1452
Jaillon O et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jena SN et al. (2012) Development and characterization of genomic and expressed SSRs for levant cotton (Gossypium herbaceum L.). Theor Appl Genet 124:565–576
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan A (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Karaoglu H, Lee CMY, Meyer W (2005) Survey of simple sequence repeats in completed fungal genomes. Mol Biol Evol 22:639–649
Kato CY, Nagai C, Moore PH, Zee F, Kim MS, Steiger DL, Ming R (2005) Intra-specific DNA polymorphism in pineapple (Ananas comosus (L.) merr.) assessed by AFLP markers. Genet Resour Crop Ev 51:815–825
Katti MV, Ranjekar PK, Gupta VS (2001) Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol 18:1161–1167
Kinsuat M, Kumar S (2007) Polymorphic microsatellite and cryptic simple repeat sequence markers in pineapples (Ananas comosus var. comosus). Mol Ecol Notes 7:1032–1035
Labbé J, Murat C, Morin E, Le Tacon F, Martin F (2011) Survey and analysis of simple sequence repeats in the Laccaria bicolor genome, with development of microsatellite markers. Curr Genet 57:75–88
Lawson MJ, Zhang L (2006) Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol 7:R14
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221
Li YC, Korol AB, Fahima T, Beiles A, Nevo E (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11:2453–2465
Li B, Xia Q, Lu C, Zhou Z, Xiang Z (2004a) Analysis on frequency and density of microsatellites in coding sequences of several eukaryotic genomes. Genomics Proteomics Bioinform 2:24–31
Li Y-C, Korol AB, Fahima T, Nevo E (2004b) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007
Li W, Feng Y, Sun H, Deng Y, Yu H, Chen H (2014) Analysis of simple sequence repeats in the Gaeumannomyces graminis var. tritici genome and the development of microsatellite markers. Curr Genet 60:237–245
Liu S-R, Li W-Y, Long D, Hu C-G, Zhang J-Z (2013) Development and characterization of genomic and expressed ssrs in citrus by genome-wide analysis. PLoS One:8. doi:10.1371/journal.pone.0075149
Luo J et al. (2012) Microsatellite mutation rate during allohexaploidization of newly resynthesized wheat. Int J Mol Sci 13:12533–12543
Luo H et al. (2015) Genome-Wide Analysis of Simple Sequence Repeats and Efficient Development of Polymorphic SSR Markers Based on Whole Genome Re-Sequencing of Multiple Isolates of the Wheat Stripe Rust Fungus. PLoS One:10. doi:10.1371/journal.pone.0130362
Mian MAR, Saha MC, Hopkins AA, Wang Z-Y (2005) Use of tall fescue EST-SSR markers in phylogenetic analysis of cool-season forage grasses. Genome 48:637–647
Ming R et al. (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47:1435–1442
Molnar SJ, Rai S, Charette M, Cober ER (2003) Simple sequence repeat (SSR) markers linked to E1, E3, E4, and E7 maturity genes in soybean. Genome 46:1024–1036
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Mrázek J, Guo X, Shah A (2007) Simple sequence repeats in prokaryotic genomes. P Natl Acad Sci 104:8472–8477
Mun J-H et al. (2006) Distribution of microsatellites in the genome of Medicago truncatula: a resource of genetic markers that integrate genetic and physical maps. Genetics 172:2541–2555
Murat C et al. (2011) Distribution and localization of microsatellites in the perigord black truffle genome and identification of new molecular markers. Fungal Genet Biol 48:592–601
Ong W, Voo CLY, Kumar SV (2012) Development of ESTs and data mining of pineapple EST-SSRs. Mol Biol Rep 39:5889–5896
Paz EY et al. (2012) Genetic diversity of Cuban pineapple germplasm assessed by AFLP Markers. Crop Breed Appl Biot 12:104–110
Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, Hamon P (2006) SSR mining in coffee tree EST databases: potential use of EST–SSRs as markers for the Coffea genus. Mol Gen Genomics 276:436–449
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Project IRGS (2005) The map-based sequence of the rice genome. Nature 436:793–800
Rodríguez D, Grajal-Martín M, Isidrón M, Petit S, Hormaza J (2013) Polymorphic microsatellite markers in pineapple (Ananas comosus (L.) Merrill). Sci Hortic-Amsterdam 156:127–130
Scaglione D, Acquadro A, Portis E, Taylor CA, Lanteri S, Knapp SJ (2009) Ontology and diversity of transcript-associated microsatellites mined from a globe artichoke EST database. BMC Genomics 10:454
Sharma AK, Ghosh I (1971) Cytotaxonomy of the family bromeliaceae. Cytologia 36:237–247
Sharma R, Gupta P, Sharma V, Sood A, Mohapatra T, Ahuja PS (2008) Evaluation of rice and sugarcane SSR markers for phylogenetic and genetic diversity analyses in bamboo. Genome 51:91–103
Shoda M et al. (2012) DNA profiling of pineapple cultivars in Japan discriminated by SSR markers. Breed Sci 62:352
Sonah H et al. (2011) Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS One:6. doi:10.1371/journal.pone.0021298
Sripaoraya S, Blackhall N, Marchant R, Power J, Lowe K, Davey M (2001) Relationships in pineapple by random amplified polymorphic DNA (RAPD) analysis. Plant Breed 120:265–267
Steele K, Price A, Shashidhar H, Witcombe J (2006) Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor Appl Genet 112:208–221
Tachida H, Iizuka M (1992) Persistence of repeated sequences that evolve by replication slippage. Genetics 131:471–478
Tangphatsornruang S et al. (2009) Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean (Vigna radiata (L.) Wilczek). BMC Plant Bio 9:137
Tautz D (1989) Hypervariabflity of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Tóth G, Gáspári Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25:1105–1111
Treco D, Arnheim N (1986) The evolutionarily conserved repetitive sequence d(TG. AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol Cell Biol 6:3934–3947
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Vásquez A, López C (2014) In silico genome comparison and distribution analysis of simple sequences repeats in cassava. Int J Genomics 2014
Wahls WP, Wallace LJ, Moore PD (1990) Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell 60:95–103
Wang J et al. (2008) Genome-wide comparative analyses of microsatellites in papaya. Trop Plant Biol 1:278–292
Wöhrmann T, Weising K (2011) In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across bromeliaceae. Theor Appl Genet 123:635–647
Yu Y, Yuan D, Liang S, Li X, Wang X, Lin Z, Zhang X (2011) Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genomics 12:15
Zdobnov EM, Apweiler R (2001) InterProScan – An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848
Zhang L et al. (2004) Preference of simple sequence repeats in coding and non-coding regions of Arabidopsis thaliana. Bioinformatics 20:1081–1086
Zhang L et al. (2006) Conservation of noncoding microsatellites in plants: implication for gene regulation. BMC Genomics 7:323
Zhao Y, Williams R, Prakash C, He G (2012) Identification and characterization of gene-based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol 12:237
Acknowledgments
This work was supported startup fund from Fujian Agriculture and Forestry University to R.M.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Paulo Arruda
Electronic supplementary material
ESM 1
(DOCX 85 kb)
Rights and permissions
About this article
Cite this article
Fang, J., Miao, C., Chen, R. et al. Genome-Wide Comparative Analysis of Microsatellites in Pineapple. Tropical Plant Biol. 9, 117–135 (2016). https://doi.org/10.1007/s12042-016-9163-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-016-9163-6