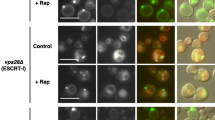
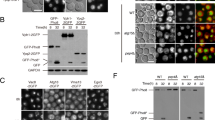
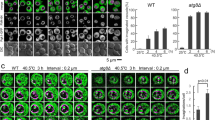
Abstract
Autophagy is a highly conserved intracellular degradation pathway in eukaryotic cells that responds to environmental changes. Genetic analyses have shown that more than 40 autophagy-related genes (ATG) are directly involved in this process in fungi. In addition to Atg proteins, most vesicle transport regulators are also essential for each step of autophagy. The present study showed that one Endoplasmic Reticulum protein in Saccharomyces cerevisiae, Tip20, which controls Golgi-to-ER retrograde transport, was also required for starvation-induced autophagy under high temperature stress. In tip20 conditional mutant yeast, the transport of Atg8 was impaired during starvation, resulting in multiple Atg8 puncta dispersed outside the vacuole that could not be transported to the pre-autophagosomal structure/phagophore assembly site (PAS). Several Atg8 puncta were trapped in ER exit sites (ERES). Moreover, the GFP-Atg8 protease protection assay indicated that Tip20 functions before autophagosome closure. Furthermore, genetic studies showed that Tip20 functions downstream of Atg5 and upstream of Atg1, Atg9 and Atg14 in the autophagy pathway. The present data show that Tip20, as a vesicle transport regulator, has novel roles in autophagy.




Similar content being viewed by others
Abbreviations
- Atg:
-
autophagy-related gene
- ER:
-
endoplasmic reticulum
- ERES:
-
endoplasmic reticulum exit sites
- GFP:
-
green fluorescent protein
- PAS:
-
pre-autophagosomal structure/phagophore assembly site
- PE:
-
phosphatidylethanolamine
- RFP:
-
red fluorescent protein.
References
Andag U, Neumann T and Schmitt HD 2001 The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem. 276 39150–39160
Cosson P, Schroder-Kohne S, Sweet DS, Demolliere C, Hennecke S, Frigerio G and Letourneur F 1997 The Sec20/Tip20p complex is involved in ER retrieval of dilysine-tagged proteins. Eur. J. Cell Biol. 73 93–97
Darsow T, Rieder SE and Emr SD 1997 A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 138 517–529
Feng Y, He D, Yao Z and Klionsky DJ 2014 The machinery of macroautophagy. Cell Res. 24 24–41
Frigerio G 1998 The Saccharomyces cerevisiae early secretion mutant tip20 is synthetic lethal with mutants in yeast coatomer and the SNARE proteins Sec22p and Ufe1p. Yeast 14 633–646
Geng J, Nair U, Yasumura-Yorimitsu K and Klionsky DJ 2010 Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 21 2257–2269
Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F and Ohsumi Y 2007 The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282 37298–37302
He C and Klionsky DJ 2007 Atg9 trafficking in autophagy-related pathways. Autophagy 3 271–274
Huang J and Klionsky DJ 2007 Autophagy and human disease. Cell Cycle 6 1837–1849
Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M and Ohsumi Y 2003 A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5 539–545
Kraynack BA, Chan A, Rosenthal E, Essid M, Umansky B, Waters MG and Schmitt HD 2005 Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol. Biol. Cell 16 3963–3977
Lemus L, Ribas JL, Sikorska N and Goder V 2016 An ER-localized SNARE protein is exported in specific COPII vesicles for autophagosome biogenesis. Cell Rep. 14 1710–1722
Lewis MJ, Rayner JC and Pelham HR 1997 A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 16 3017–3024
Lipatova Z, Majumdar U and Segev N 2016 Trs33-Containing TRAPP IV: A Novel Autophagy-Specific Ypt1 GEF. Genetics 204 1117–1128
Liu X, Mao K, Yu AY, Omairi-Nasser A, Austin J, 2nd, Glick BS, Yip CK and Klionsky DJ 2016 The Atg17-Atg31-Atg29 Complex coordinates with Atg11 to recruit the Vam7 SNARE and mediate autophagosome-vacuole fusion. Curr. Biol. 26 150–160
Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S and Klionsky DJ 2010 Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 107 7811–7816
Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ and Reggiori F 2010 An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190 1005–1022
Meiringer CT, Rethmeier R, Auffarth K, Wilson J, Perz A, Barlowe C, Schmitt HD and Ungermann C 2011 The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors (SNAREs): implications for fusion and fusion regulation. J. Biol. Chem. 286 25039–25046
Mizushima N, Levine B, Cuervo AM and Klionsky DJ 2008 Autophagy fights disease through cellular self-digestion. Nature 451 1069–1075
Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew JA, Jiang X, Reggiori F, Melia TJ and Klionsky DJ 2011a SNARE proteins are required for macroautophagy. Cell 146 290–302
Nair U, Thumm M, Klionsky DJ and Krick R 2011b GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy 7 1546–1550
Nakatogawa H, Ichimura Y and Ohsumi Y 2007 Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130 165–178
Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y and Klionsky DJ 2000 Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148 465–480.
Obara K, Sekito T and Ohsumi Y 2006 Assortment of phosphatidylinositol 3-kinase complexes–Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell 17 1527–1539
Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR and Rubinsztein DC 2010 Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 90 1383–1435
Reggiori F, Shintani T, Nair U and Klionsky DJ 2005 Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 1 101–109
Reggiori F, Tucker KA, Stromhaug PE and Klionsky DJ 2004 The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6 79–90
Suzuki K, Kubota Y, Sekito T and Ohsumi Y 2007 Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12 209–218
Sweet DJ and Pelham HR 1993 The TIP1 gene of Saccharomyces cerevisiae encodes an 80 kDa cytoplasmic protein that interacts with the cytoplasmic domain of Sec20p. EMBO J. 12 2831–2840
Vida TA and Emr SD 1995 A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128 779–792
Wang J, Davis S, Menon S, Zhang J, Ding J, Cervantes S, Miller E, Jiang Y and Ferro-Novick S 2015 Ypt1/Rab1 regulates Hrr25/CK1delta kinase activity in ER-Golgi traffic and macroautophagy. J. Cell Biol. 210 273–285
Xie Z and Klionsky DJ 2007 Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9 1102–1109
Yang S and Rosenwald AG 2016 Autophagy in Saccharomyces cerevisiae requires the monomeric GTP-binding proteins, Arl1 and Ypt6. Autophagy 12 1721–1737
Yang Z and Klionsky DJ 2010 Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12 814–822.
Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M and Klionsky DJ 2010 The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188 101–114
Zink S, Wenzel D, Wurm CA and Schmitt HD 2009 A link between ER tethering and COP-I vesicle uncoating. Dev. Cell 17 403–416
Zou S, Sun D and Liang Y 2017 The Roles of the SNARE protein Sed5 in autophagy in Saccharomyces cerevisiae. Mol. Cell. 40 643–654
Acknowledgements
We are grateful to H. Schmitt for providing WT, tip20-5 and dsl1-22 strains, J Nunnaria for providing DsRed-HDEL-305 plasmids, and Q Jin for providing pFA6a-tdTomato-KanMx plasmid. This work was supported by grants from the Natural Science Foundation of China (31301173 to SZ and 31101428 to LC) and Shandong Agricultural University Talent Introduction Funding (20171226 to HD).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Sudha Bhattacharya
Rights and permissions
About this article
Cite this article
Chen, L., Zhang, C., Liang, Y. et al. Autophagy requires Tip20 in Saccharomyces cerevisiae. J Biosci 44, 17 (2019). https://doi.org/10.1007/s12038-018-9839-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-018-9839-1