Abstract
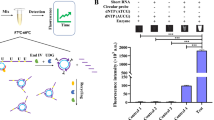
Neisseria gonorrhoeae (NG) is the second most common bacterial sexually transmitted infection (STI) worldwide. Gonorrhoea is a very serious infection because if untreated, it can lead to significant ramifications to reproductive, maternal, & newborn health and increase transmission of HIV. Infections are very often asymptomatic and symptoms when present manifest differently in men and women. The cornerstone of gonorrhoea control is to assure rapid diagnosis and prompt treatment of patients to prevent the onward spread of infection. The resource-rich settings are utilizing nucleic acid amplification tests (NAATs) for diagnosis, whereas resource-limited settings like ours where laboratory infrastructure is lacking, reliance is placed on syndromic approach. In view of the limitations of each, there is a compelling need for development of a point of care test (POCT). Aptamers offer such potential. These are short oligonucleotides that bind to its target with high affinity and specificity and therefore can be maneuvred for use in diagnostics. In this study, we performed live cell-SELEX (Systematic Evolution of Ligands by EXponential enrichment) to select 12 single-stranded DNA (ssDNA) aptamers that bind strongly to a cocktail of Neisseria gonorrhoeae strains, with Kd values ranging from 8.58 to 596 nM. Gold nanoparticle (GNP) assay revealed that one of the aptamers, E8 19 was highly specific for Neisseria gonorrhoeae (Kd = 24.5 nM). More importantly, it did not demonstrate any binding to Neisseria meningitidis and commensal Neisseria sp. The identified aptamer holds much promise for the development of a rapid test for diagnosis of gonorrhoea.


Similar content being viewed by others
References
Web Annex 1. Key data at a glance. In: Global progress report on HIV, viral hepatitis and sexually transmitted infections, 2021. Accountability for the global health sector strategies 2016–2021: actions for impact. Geneva: World Health Organization; 2021. Licence: CC BY-NC-SA 3.0 IGO.
Verma, R., Sood, S., Singh, R., et al. (2014). Coupling electrochemical response of a DNA biosensor with PCR for Neisseria gonorrhoeae detection. Diagnostic Microbiology and Infectious Disease, 78(1), 16–23. https://doi.org/10.1016/j.diagmicrobio.2013.09.010
Meyer, T., & Buder, S. (2020). The laboratory diagnosis of Neisseria gonorrhoeae: Current testing and future demands. Pathogens, 9(2), 91. https://doi.org/10.3390/pathogens9020091
Morris, K. N., Jensen, K. B., Julin, C. M., et al. (1998). High affinity ligands from in vitro selection: Complex targets. ProcNatlAcadSci USA, 95, 2902–2907.
Homann, M., & Goringer, H. U. (1999). Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Research, 27, 2006–2014.
Cao, X., Li, S., Chen, L., et al. (2009). Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Research, 37, 4621–4628.
Dwivedi, H. P., Smiley, R. D., & Jaykus, L. A. (2010). Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Applied Microbiology and Biotechnology, 87, 2323–2334.
Lee, Y. J., Han, S. R., Maeng, J. S., et al. (2012). In vitro selection of Escherichia coli O157:H7- specific RNA aptamer. BiochemBiophysical Research Communication, 417, 414–420.
Wu, W. H., Li, M., Wang, Y., et al. (2010). Aptasensors for rapid detection of Escherichia coli O157:H7 and Salmonella typhimurium. Nanoscale Research Letters, 7(658–664), 20.
Yuan, J. L., Tao, Z., Yu, Y., et al. (2014). A visual detection method for Salmonella typhimurium based on aptamer recognition and nanogold labeling. Food Control, 37, 188–192.
Xiong, L., Xia, M., Wang, Q., Meng, Z., Zhang, J., Yu, G., Dong, Z., Lu, Y., & Sun, Y. (2022). DNA aptamers specific for Legionella pneumophila: Systematic evolution of ligands by exponential enrichment in whole bacterial cells. Biotechnology Letters, 44(5–6), 777–786. https://doi.org/10.1007/s10529-022-03252-z
Kim, Y. S., Kim, J. H., Kim, I. A., et al. (2010). A novel colorimetric aptasensor using gold nanoparticle for a highly sensitive and specific detection of oxytetracycline. Biosensors & Bioelectronics, 26(4), 1644–1649.
Samarawickrama, A., Alexander, S., & Ison, C. (2011). A laboratory-based evaluation of the BioStar optical IMMUNOASSAY point-of-care test for diagnosing Neisseria gonorrhoeae infection. J Medicine Microbiology, 60(12), 1779–1781.
Rahman, M. S., Beever, W., Skov, S., et al. (2013). Using urinary leucocyte esterase tests as an indicator of infection with gonorrhoea or chlamydia in asymptomatic males in a primary health care setting. International Journal of STD and AIDS, 25(2), 138–144.
Abbai, N. S., Moodley, P., et al. (2015). Clinical evaluation of the onestep gonorrhea rapicard instatest for detection of Neisseria gonorrhoeae in symptomatic patients from KwaZulu-Natal. South African Journal Clinic Microbiology, 53, 1348–1350.
Adamson, P. C., Loeffelholz, M. J., & Klausner, J. D. (2020). Point-of-Care testing for sexually transmitted infections a review of recent developments. Archives of Pathology and Laboratory Medicine, 144(11), 1344–1351. https://doi.org/10.5858/arpa.2020-0118-RA
Marton, S., Cleto, F., Krieger, M. A., et al. (2016). Isolation of an aptamer that binds specifically to E. coli. PLoS ONE, 11(4), e0153637. https://doi.org/10.1371/journal.pone.0153637
Malhotra, S., Pandey, A., Rajput, Y., et al. (2014). Selection of aptamers for aflatoxin M1 and their characterization. Journal Molecular Recognition, 27(8), 493–500.
Musheev, M. U., & Krylov, S. N. (2006). Selection of aptamers by systematic evolution of ligands by exponential enrichment: Addressing the polymerase chain reaction issue. Analytica Chimica Acta, 564, 91–96.
Mayer, G., Ahmed, M. S., Dolf, A., Endl, E., Knolle, P. A., & Famulok, M. (2010). Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nature Protocols, 5(12), 1993–2004. https://doi.org/10.1038/nprot.2010.163
Savory, N., Nzakizwanayo, J., Abe, K., et al. (2014). Selection of DNA aptamers against uropathogenic Escherichia coli NSM59 by quantitative PCR controlled Cell-SELEX. Journal of Microbiol Methods, 104, 94–100. https://doi.org/10.1016/j.mimet.2014.06.016
Duan, N., Ding, X., Wu, S., Xia, Y., Ma, X., Wang, Z., & Chen, J. (2013). In vitro selection of a DNA aptamer targeted against Shigella dysenteriae. Journal of Microbiol Methods, 94, 170–174. https://doi.org/10.1016/j.mimet.2013.06.016
Mozioglu, E., Gokmen, O., Tamerler, C., et al. (2016). Selection of Nucleic acid aptamers specific for Mycobacterium tuberculosis. Applied Biochemistry and Biotechnology, 178, 849–864. https://doi.org/10.1007/s12010-015-1913-7
Ledlod, S., Areekit, S., Santiwatanakul, S., et al. (2020). Colorimetric aptasensor for detecting salmonella spp., listeria monocytogenes, and E. coli in meat samples. Food Science Technology International, 26(5), 430–443.
Chavez, J. L., Hagen, J. A., & Kelley-Loughnane, N. (2017). Fast and selective plasmonic serotonin detection with aptamer-gold nanoparticle conjugates. Sensors, 17, 681. https://doi.org/10.3390/s17040681
Bakhtiari, H., Palizban, A. A., Khanahmad, H., & Mofid, M. R. (2021). Novel approach to overcome defects of cell-SELEX in developing aptamers against Aspartate β-hydroxylase. ACS Omega, 6(16), 11005–11014. https://doi.org/10.1021/acsomega.1c00876
Acknowledgements
The work was supported by funding from Department of Health Research, Ministry of Health & Family Welfare, India (Grant No. DHR/HRD/WS-03-2015-16). The authors gratefully acknowledge Professor Monica M Lahra & Dr. Athena Limnios, Neisseria Reference Laboratory and WHO Collaborating Center for STD, SEALS Microbiology, The Prince of Wales Hospital, Australia for presenting commensal Neisseria sp. to our laboratory. We are also thankful to Dr. Benu Dhawan for providing Mycoplasma hominis NCTC 10111 and Ureaplasma urealyticum NCTC 10177. The authors acknowledge Shri Rajendra Singh for his technical assistance.
Funding
The work was supported by funding from Department of Health Research, Ministry of Health & Family Welfare, India (Grant No. DHR/HRD/WS-03–2015-16).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. SM: Carried out the research work. The first draft of the manuscript was written by SM and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest to disclose.
Ethical Approval
Not Applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Malhotra, S., Gupta, S. & Sood, S. Selection of DNA Aptamers Against Neisseria gonorrhoeae Causing Sexually Transmitted Infection (STI). Mol Biotechnol 65, 2099–2107 (2023). https://doi.org/10.1007/s12033-023-00688-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00688-0